The Observational Health Data Sciences and Informatics (OHDSI) international community hosted a COVID-19 virtual study-a-thon March 26-29 to inform healthcare decision-making in response to the current global pandemic. That work served as the foundation for global community research into characterization, population-level effect estimation and patient-level prediction questions about the disease. Ongoing work continues in the OHDSI MSTeams environment; if you wish to join the OHDSI Teams environment, please fill this form out. Once there, you can tell us which working groups or studies you would like to join.
Published Studies
Risk of hydroxychloroquine alone and in combination with azithromycin in the treatment of rheumatoid arthritis: a multinational, retrospective study. Lane JCE, Weaver J, Kostka K, Duarte-Salles T, Abrahao MTF, Alghoul H, Alser O, Alshammari TM, Biedermann P, Banda JM, Burn E, Casajust P, Conover MM, Culhane AC, Davydov A, DuVall SL, Dymshyts D, Fernandez-Bertolin S, Fišter K, Hardin J, Hester L, Hripcsak G, Kaas-Hansen BS, Kent S, Khosla S, Kolovos S, Lambert CG, van der Lei J, Lynch KE, Makadia R, Margulis AV, Matheny ME, Mehta P, Morales DR, Morgan-Stewart H, Mosseveld M, Newby D, Nyberg F, Ostropolets A, Park RW, Prats-Uribe A, Rao GA, Reich C, Reps J, Rijnbeek P, Sathappan SMK, Schuemie M, Seager S, Sena AG, Shoaibi A, Spotnitz M, Suchard MA, Torre CO, Vizcaya D, Wen H, de Wilde M, Xie J, You SC, Zhang L, Zhuk O, Ryan P, Prieto- Alhambra D; OHDSI-COVID-19 consortium. Lancet Rheumatol. 2020 Nov;2(11):e698-e711. doi: 10.1016/S2665-9913(20)30276-9. Epub 2020 Aug 21. PMID: 32864627; PMCID: PMC7442425. [OHDSI press release]
Deep phenotyping of 34,128 adult patients hospitalised with COVID-19 in an international network study. Burn E, You SC, Sena AG, Kostka K, Abedtash H, Abrahão MTF, Alberga A, Alghoul H, Alser O, Alshammari TM, Aragon M, Areia C, Banda JM, Cho J, Culhane AC, Davydov A, DeFalco FJ, Duarte-Salles T, DuVall S, Falconer T, Fernandez- Bertolin S, Gao W, Golozar A, Hardin J, Hripcsak G, Huser V, Jeon H, Jing Y, Jung CY, Kaas-Hansen BS, Kaduk D, Kent S, Kim Y, Kolovos S, Lane JCE, Lee H, Lynch KE, Makadia R, Matheny ME, Mehta PP, Morales DR, Natarajan K, Nyberg F, Ostropolets A, Park RW, Park J, Posada JD, Prats-Uribe A, Rao G, Reich C, Rho Y, Rijnbeek P, Schilling LM, Schuemie M, Shah NH, Shoaibi A, Song S, Spotnitz M, Suchard MA, Swerdel JN, Vizcaya D, Volpe S, Wen H, Williams AE, Yimer BB, Zhang L, Zhuk O, Prieto-Alhambra D, Ryan P. Nat Commun. 2020 Oct 6;11(1):5009. doi: 10.1038/s41467-020-18849-z. PMID: 33024121; PMCID: PMC7538555. [OHDSI press release]
Renin-angiotensin system blockers and susceptibility to COVID-19: an international, open science, cohort analysis. Morales DR, Conover MM, You SC, Pratt N, Kostka K, Duarte-Salles T, Fernández-Bertolín S, Aragón M, DuVall SL, Lynch K, Falconer T, van Bochove K, Sung C, Matheny ME, Lambert CG, Nyberg F, Alshammari TM, Williams AE, Park RW, Weaver J, Sena AG, Schuemie MJ, Rijnbeek PR, Williams RD, Lane JCE, Prats-Uribe A, Zhang L, Areia C, Krumholz HM, Prieto-Alhambra D, Ryan PB, Hripcsak G, Suchard MA. Lancet Digit Health. 2021 Feb;3(2):e98-e114. doi: 10.1016/S2589-7500(20)30289-2. Epub 2020 Dec 17. PMID: 33342753; PMCID: PMC7834915. [OHDSI press release]
Risk of depression, suicide and psychosis with hydroxychloroquine treatment for rheumatoid arthritis: a multinational network cohort study. Lane JCE, Weaver J, Kostka K, Duarte-Salles T, Abrahao MTF, Alghoul H, Alser O, Alshammari TM, Areia C, Biedermann P, Banda JM, Burn E, Casajust P, Fister K, Hardin J, Hester L, Hripcsak G, Kaas-Hansen BS, Khosla S, Kolovos S, Lynch KE, Makadia R, Mehta PP, Morales DR, Morgan-Stewart H, Mosseveld M, Newby D, Nyberg F, Ostropolets A, Woong Park R, Prats-Uribe A, Rao GA, Reich C, Rijnbeek P, Sena AG, Shoaibi A, Spotnitz M, Vignesh S, Suchard MA, Vizcaya D, Wen H, de Wilde M, Xie J, You SC, Zhang L, Lovestone S, Ryan P, Prieto-Alhambra D; OHDSI-COVID-19 consortium. Rheumatology (Oxford). 2020 Dec 25:keaa771. doi: 10.1093/rheumatology/keaa771. Epub ahead of print. PMID: 33367863; PMCID: PMC7798671.
Can we trust the prediction model? Illustrating the importance of external validation by implementing the COVID-19 Vulnerability (C-19) Index across an international network of observational healthcare datasets. Reps JM, Kim C, Williams RD, Markus AF, Yang C, Salles TD, Falconer T, Jonnagaddala J, Williams A, Fernández-Bertolín S, DuVall SL, Kostka K, Rao G, Shoaibi A, Ostropolets A, Spotnitz ME, Zhang L, Casajust P, Steyerberg EW, Nyberg F, Kaas-Hansen BS, Choi YH, Morales D, Liaw ST, Abrahão MTF, Areia C, Matheny ME, Aragón M, Park RW, Hripcsak G, Reich CG, Suchard MA, You SC, Ryan PB, Prieto-Alhambra D, Rijnbeek PR. JMIR Med Inform. 2021 Feb 27. doi: 10.2196/21547. MID: 33661754.
Characteristics, outcomes, and mortality amongst 133,589 patients with prevalent autoimmune diseases diagnosed with, and 48,418 hospitalised for COVID-19: a multinational distributed network cohort analysis. Tan EH, Sena AG, Prats-Uribe A, You SC, Ahmed WU, Kostka K, Reich C, Duvall SL, Lynch KE,Matheny ME, Duarte-Salles T, Bertolin SF, Hripcsak G, Natarajan K, Falconer T, Spotnitz M, Ostropolets A, Blacketer C, Alshammari TM, Alghoul H, Alser O, Lane JCE, Dawoud DM, Shah K, Yang Y, Zhang L, Areia C, Golozar A, Relcade M, Casajust P, Jonnagaddala J, Subbian V, Vizcaya D, Lai LY, Nyberg F, Morales DR, Posada JD, Shah NH, Gong M, Vivekanantham A, Abend A, Minty EP, Suchard M, Rijnbeek P, Ryan PB, Prieto-Alhambra D. Rheumatology (Oxford). 2021 Mar 16:keab250. doi: 10.1093/rheumatology/keab250. PMID: 33269355; PMCID: PMC7709171.
Comparative Effectiveness of Famotidine in Hospitalized COVID-19 Patients. The American Journal of Gastroenterology. Shoaibi A, Fortin S, Weinstein R, Berlin J, Ryan, P. 2021 Jan 28. doi: 10.14309/ajg.0000000000001153
Use of repurposed and adjuvant drugs in hospital patients with covid-19: multinational network cohort study. Prats-Uribe A, Sena AG, Lai LY, Ahmed W, Alghoul H, Alser O, Alshammari TM, Areia C, Carter W, Casajust P, Dawoud D, Golozar A, Jonnagaddala J, Mehta P, Gong M, Morales DR, Nyberg F, Posada J, Recalde M, Roel E, Shah K, Shah NH, Schilling L, Subbian V, Vizcaya D, Zhang L, Zhang Y, Zhu H, Liu L, Cho J, You SC, Rijnbeek P, Hripcsak G, Lane JCE, Burn E, Reich C, Suchard M, Duarte-Salles T, Kostka K, Ryan PB, Prieto-Alhambra D. BMJ. 373 2021; doi:10.1136/bmj.n1038. [OHDSI press release]
30-day outcomes of children and adolescents with COVID-19: an international experience. Duarte-Salles T, Vizcaya D, Pistillo A, Casajust P, Sena AG, Lai LY, Prats-Uribe A, Ahmed W, Alshammari TM, Alghoul H, Alser O, Burn E, You SC, Areia C, Blacketer C, Duvall S, Falconer T, Carter W, Fernandez-Bertolin S, Fortin S, Golozar A, Gong M, Tan EH, Huser V, Iveli P, Morales DR, Nyberg F, Posada J, Recalde M, Roel E, Schilling L, Shah NH, Shah K, Suchard M, Zhang L, Zhang Y, Williams A, Reich C, Hripcsak G, Rijnbeek P, Ryan PB, Kostka K, Prieto-Alhambra D. Pediatrics. 2021; doi: 10.1542/peds.2020-042929. [OHDSI press release]
Characterising the background incidence rates of adverse events of special interest for covid-19 vaccines in eight countries: multinational network cohort study. Li X, Ostropolets A, Makadia R, Shoaibi A, Rao G, Sena AG, Martinez-Hernandez E, Delmestri A, Verhamme K, Rijnbeek P, Duarte-Salles T, Suchard M, Ryan PB, Hripcsak G, Prieto-Alhambra D. BMJ 2021;373:n1435; doi: 10.1136/bmj.n1435. [OHDSI press release]
Characteristics and outcomes of 627 044 COVID-19 patients living with and without obesity in the United States, Spain, and the United Kingdom. Recalde M, Roel E, Pistillo A, Sena AG, Prats-Uribe A, Ahmed WU, Alghoul H, Alshammari TM, Alser O, Areia C, Burn E, Casajust P, Dawoud D, DuVall SL, Falconer T, Fernández-Bertolín S, Golozar A, Gong M, Lai LYH, Lane JCE, Lynch KE, Matheny ME, Mehta PP, Morales DR, Natarjan K, Nyberg F, Posada JD, Reich CG, Rijnbeek PR, Schilling LM, Shah K, Shah NH, Subbian V, Zhang L, Zhu H, Ryan P, Prieto-Alhambra D, Kostka K, Duarte-Salles T. Int J Obes (Lond). 2021 Jul 15. doi: 10.1038/s41366-021-00893-4.
Preprints
Prieto-Alhambra D, et al. Unraveling COVID-19: a large-scale characterization of 4.5 million COVID-19 cases using CHARYBDIS. doi: 10.21203/rs.3.rs-279400/v1
Williams R, et al. Seek COVER: Development and validation of a personalized risk calculator for COVID-19 outcomes in an international network. doi: 10.1101/2020.05.26.20112649
Lai L, et al. Clinical characteristics, symptoms, management and health outcomes in 8,598 pregnant women diagnosed with COVID-19 compared to 27,510 with seasonal influenza in France, Spain and the US: a network cohort analysis. doi: 10.1101/2020.10.13.20211821
Kohler J, et al. Using Real World Data to Understand HIV and COVID-19 in the U.S.A. and Spain: Characterizing Co-Infected Patients Across the Care Cascade. doi: 10.1101/2020.11.10.20229401
Golozar A, et al. Baseline phenotype and 30-day outcomes of people tested for COVID-19: an international network cohort including >3.32 million people tested with real-time PCR and >219,000 tested positive for SARS-CoV-2 in South Korea, Spain and the United States. doi: 10.1101/2020.10.25.20218875
Burn E, et al. Use of dialysis, tracheostomy, and extracorporeal membrane oxygenation among 842,928 patients hospitalized with COVID-19 in the United States. doi: 10.1101/2020.11.25.20229088
Nishimuri A, et al. Alpha-1 blockers and susceptibility to COVID-19 in benign prostate hyperplasia patients: an international cohort study. doi: 10.1101/2021.03.18.21253778
Study-A-Thon Links
• 88 Hours (Study-a-thon feature)
• OHDSICOVID19 Closing Call
Updates
(Future COVID-19 Updates Will Be In News/Updates Section and OHDSI Social Channels)
September 3
 “Characteristics and outcomes of 627,044 COVID-19 patients with and without obesity in the United States, Spain, and the United Kingdom,” a new OHDSI study generated by the CHARYBDIS team, is available as a preprint on MedRxiv.
“Characteristics and outcomes of 627,044 COVID-19 patients with and without obesity in the United States, Spain, and the United Kingdom,” a new OHDSI study generated by the CHARYBDIS team, is available as a preprint on MedRxiv.
The prevalence of obesity in 627,044 COVID-19 patients was more common than in the 4.5 million influenza patients used in the study, which spanned six databases. Hospitalization, intensive services, and fatality rates were higher within obese COVID-19 patients than non-obese ones.
The prevalence of obesity was higher among hospitalized COVID-19 (range: 38% to 54%) than either diagnosed COVID-19 (30% to 47%) or diagnosed/hospitalized influenza (15% to 48%) patients. Obese hospitalized COVID-19 patients were more often female and younger than non-obese COVID-19 patients or obese influenza patients.
Obese COVID-19 patients were also more likely to have prior comorbidities, present with cardiovascular and respiratory events during hospitalization, require intensive services, or die compared to non-obese COVID-19 patients.
The research team, led by lead authors Martina Recalde and Elena Roel, hopes these findings can guide preventative strategies for infection/complications. The research team is seeking feedback on the study, which is currently under the peer-review process.
The CHARYBDIS (Characterizing Health Associated Risks, and Your Baseline Disease In SARS-COV-2) Project is an OHDSI initiative that attempts to describe the baseline demographic, clinical characteristics, treatments and outcomes of interest among individuals tested for SARS-CoV-2 and/or diagnosed with COVID-19 overall and stratified by sex, age and specific comorbidities, as compared to Influenza patients from 2017-2018. More information on CHARYBDIS is available on its GitHub page.
Preprints are preliminary reports of work that have not been certified by peer review. They should not be relied on to guide clinical practice or health-related behavior and should not be reported in news media as established information.
August 22
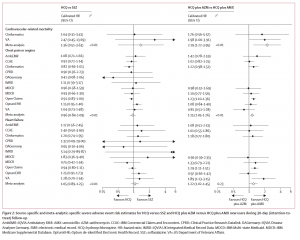
Source-specific and meta-analytic-specific severe adverse event risk estimates for HCQ versus SSZ and HCQ plus AZM versus HCQ plus AMX new users during 30-day (intention-to-treat)
follow-up.
The combination of hydroxychloroquine (HCQ) and azithromycin (AZM) has been linked to significant cardiovascular risks, including mortality, in the largest safety study ever performed on both HCQ and HCQ+AZM. This network study, led by the Observational Health Data Sciences and Informatics community, was recently published in Lancet Rheumatology.
In patients with rheumatoid arthritis, HCQ treatment in the short term (30 days) was found to not carry an excess risk of complications associated with its use, but HCQ treatment in the long term had a 65% relative increase in cardiovascular-related mortality, compared to sulfasalazine.
HCQ + AZM had a cardiovascular mortality risk that was more than twice (2.19) as high as the comparative treatment even in the short term based on findings from more than 320,000 users of that combination therapy. This treatment also produced a 15-20% increased rate of angina/chest pain and heart failure.
The full paper is available here.
This study, first released on MedRxiv, has already made significant impacts in the healthcare community. On April 23, the European Medicines Agency (EMA) cited the study in a warning about the risk of serious side effects with chloroquine and hydroxychloroquine. In July, the EMA again highlighted the study, among other efforts within the OHDSI community, in its eighth revision of The European Network of Centres for Pharmacoepidemiology and Pharmacovigilance (ENCePP) Guide on Methodological Standards in Pharmacoepidemiology.
This is the first published study to be generated from the OHDSI COVID Study-a-thon, a global effort in March to set the foundation for OHDSI efforts to design and execute network observational studies around characterization, patient-level prediction and population-level effect estimation to inform decision-making around the global pandemic. Multiple studies, several of which are highlighted later, have been posted to MedRxiv and are currently under peer review.
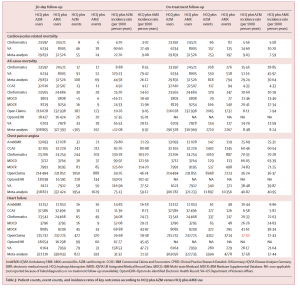
Patient counts, event counts, and incidence rates of key outcomes according to HCQ plus AZM versus HCQ plus AMX use
“Hydroxychloroquine, both alone and in combination with azithromycin, gained strong consideration as a potential COVID treatment without a large-scale study of its overall safety profile,” said Daniel Prieto-Alhambra, MD, MSc, PhD, co-senior author on this study. “We had access to an unprecedented amount of data on this drug, and we were relieved to find no worrying side effects in the short-term use of hydroxychloroquine. However, when prescribed in combination with azithromycin, it may induce heart failure and cardiovascular mortality and we would urge caution in using the two together.”HCQ, a drug commonly used in the treatment of malaria, lupus and rheumatoid arthritis (RA), gained early attention during the pandemic as a potential COVID-19 treatment. The short-term (<30 days) safety profile did not identify excess risk in any of 16 severe adverse events as compared to a similar RA drug, sulfasalazine (SSZ). Long-term HCQ therapy was associated with a 65% increase in cardiovascular mortality as compared to SSZ.
This study examined more than 950,000 HCQ users through deidentified electronic health records and administrative claims data over a 20-year period. Records were collected from 14 different databases spanning six nations (Germany, Japan, Netherlands, Spain, United Kingdom, United States) and then mapped to the OMOP Common Data Model to generate this large-scale analysis.
“At medical school we were taught to ‘first do no harm’ and to me, our study focuses on this core belief of modern medicine,” said Jennifer Lane, MD, who served as co-lead author on this study along with Jamie Weaver, MPH, MS. “OHDSI has the power to investigate this question in a very thorough way and to go through rigorous steps. We are looking at patients from the general population, which is why it is so important to look at data from multiple countries. There are reasons why you may get bias from one data source, but if we find a signal in the Netherlands, and we find it in Spain, and we find it in the U.S., then we know we have something.”
The study was developed and executed by the OHDSI (Observational Health Data Sciences and Informatics) community, a multi-stakeholder, interdisciplinary collaborative to bring out the value of health data through large-scale analytics. All solutions are open-source, and links to the study protocol, code and results are posted at the bottom of this release.
“It required a global effort to generate this level of reproducible, reliable real-world evidence to inform decision-making around COVID treatment,” said Patrick Ryan, PhD, co-senior author on this study. “Our community collaborated for years to develop the high-level analytics which set the course for these studies. Standardizing data for nearly 1,000,000 patients on hydroxychloroquine provides confidence in these findings, and we are pleased to see that this study has already helped make a positive clinical impact as treatment options continue to be evaluated.”
August 8
Following concern in the rheumatological community regarding recent regulatory warnings that hydroxychloroquine used in the COVID-19 pandemic could cause acute psychiatric events, the OHDSI network completed a study that is currently under peer review and posted to MedRxiv, entitled “Risk of depression, suicidal ideation, suicide and psychosis with hydroxychloroquine treatment for rheumatoid arthritis: a multi-national network cohort study.”
A total of 918,144 and 290,383 users of hydroxychloroquine and sulfasalazine, respectively, were included from 10 sources and 3 countries (Germany, UK and US) and study outcomes included depression, suicide/suicidal ideation, and hospitalization for psychosis.
The study concluded that hydroxychloroquine as used to treat RA does not appear to increase the risk of depression, suicide/suicidal ideation, or psychosis compared to sulfasalazine. No effects were seen in the short or long term. Use at a higher dose or for different indications needs further investigation.
July 16
• OHDSI was heavily represented once again during the weekly FDA/Reagan Udall COVID-19 Evidence Accelerator meeting. First, Patrick Ryan presented on behalf of the OHDSI COVID-19 team on CHARYBDIS. Afterward, Juan Banda presented on his collaborative work with Daniel Prieto-Alhambra on Twitter data. Slides to both presentations are available here.
• Study leads from the Characterization team met Tuesday to provide updates on progress on the multiple studies being generated from CHARYBDIS. Several of those updates are highlighted in the chat section of the Study-Characterization channel. Study leads will be meeting weekly (Tuesday, 11 am ET/4 pm UK/5 pm CET) to continue these discussions. We are excited about the important manuscripts that can come from this exciting work.
• CHARYBDIS updates continue to be updated here: https://data.ohdsi.org/Covid19CharacterizationCharybdis/. There are now 13 databases from three different continents included in the study. Thank you to all partners for your work to help build our data network so we can generate robust, reliable evidence. We continue to seek new partners, so if you are interested in joining our efforts, please contact Talita Duarte-Salles, Kristin Kostka, or Albert Prats-Uribe.
• Today is the last call for abstracts for the 2020 OHDSI Symposium Collaborator Showcase. While our showcase is not limited to COVID-19 work, we know that there have been many important studies generated around this pandemic, so please remember to share your abstracts before tonight’s 8 pm ET deadline. For more information, visit the Collaborator Showcase page.
June 30
• Results from our CHARYBDIS Project continue to build, thanks in large part to the ongoing efforts of Anthony Sena, Talita Duarte-Salles, Kristin Kostka, and Albert Prats-Uribe. You can evaluate the updated results here, which now include seven databases from around the world (IPCI, SIDIAP, CUIMC, HM, PREMIER, CPRD, DEID). We know many collaborators in the community are excited to work on various studies within CHARYBDIS, so please visit the Teams environment as soon as possible so you can connect with collaborators and contribute to the many important manuscripts that can come out of this exciting work.
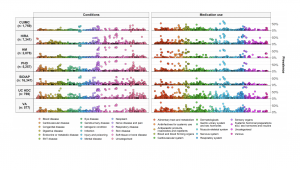
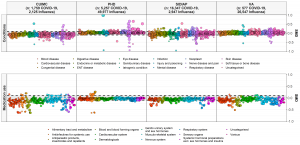 • As you can see from the MedRxiv links above, our original Characterization study “Deep phenotyping of 34,128 patients hospitalized with COVID-19 and a comparison with 81,596 influenza patients in America, Europe, and Asia: an international network study” has been updated as of June 28. The study now includes about 27,000 more COVID patients and has several graphics (including one on the right) to go along with this important manuscript that is under peer review.
• As you can see from the MedRxiv links above, our original Characterization study “Deep phenotyping of 34,128 patients hospitalized with COVID-19 and a comparison with 81,596 influenza patients in America, Europe, and Asia: an international network study” has been updated as of June 28. The study now includes about 27,000 more COVID patients and has several graphics (including one on the right) to go along with this important manuscript that is under peer review.
June 17
• Project CHARYBDIS is being run over several databases, an exciting development in this important community effort. Preliminary results from five databases (IPCI, TRDW (Tufts), HM, PREMIER, CPRD) are available here, and we continue to seek other data partners to run this study. The protocol and study code is available here: https://github.com/ohdsi-studies/Covid19CharacterizationCharybdis
• There is a working manuscript from our estimation team currently entitled “Risk of depression, suicidal ideation, suicide and psychosis with hydroxychloroquine treatment for rheumatoid arthritis: a multi-national network cohort study” and located in the Study-Estimation-Hydroxychloroquine channel. The authors are looking for community feedback, as well as ICJME forms for anybody who would like to be included as an author on the study.
• Congrats to lead author Jenna Reps and the prediction team on another MedRxiv preprint: Can we trust the prediction model? Demonstrating the importance of external validation by investigating the COVID-19 Vulnerability (C-19) Index across an international network of observational healthcare datasets. You can read the preprint here. External validation is critical for any prediction model, and through OHDSI analytics and global collaboration, this study suggests that C-19 should not be used to aid decision making during the COVID-19 pandemic.
June 12
 The Observational Health Data Sciences and Informatics (OHDSI) collaboration released a preprint entitled “Renin-angiotensin system blockers and susceptibility to COVID-19: a multinational open science cohort study.”
The Observational Health Data Sciences and Informatics (OHDSI) collaboration released a preprint entitled “Renin-angiotensin system blockers and susceptibility to COVID-19: a multinational open science cohort study.”
Preprints are preliminary reports of work that have not been certified by peer review. They should not be relied on to guide clinical practice or health-related behavior and should not be reported in news media as established information.
The preprint is available here.
There was no clear increased risk of COVID-19 diagnosis, hospitalization, or subsequent complications found for users of either angiotensin-converting enzyme inhibitors (ACEs) or angiotensin receptor blockers (ARBs) among a multinational cohort of more than 1.1 million patients using antihypertensives.
This study, the most comprehensive one to date of COVID-19 susceptibility risks for antihypertensive users, examined electronic health records from a trio of data sources from the United States and Spain (Columbia University Irving Medical Center, the Department of Veteran Affairs and SIDIAP) to conduct a systematic cohort study of ACE, ARB, calcium channel blocker (CCB) and thiazide diuretic (THZ) users.
“We currently lack reliable, transparent and generalizable evidence to inform antihypertensive choice in light of COVID-19,” says Dr. Marc Suchard, a professor at UCLA and research team leader. “This work strives to openly and reproducibly leverage real-world data to help.”
As a consequence, the study, powered by open-source tools and global collaboration within the OHDSI community, reinforces current clinical guidelines surrounding antihypertensive therapy. The findings indicate that patients should continue their ACE or ARB therapy, despite early concerns about potential risks.
Furthermore, the findings showed no clinical reason to switch from an ARB to ACE to minimize COVID-19 risk. “Based on our results, if there is a risk difference, it’s marginal and would be very challenging to further refine outside such a large-scale international study,” Dr. Suchard says.
The International COVID-ACE Receptor Inhibition Utilization and Safety (ICARIUS) protocol, code, and results are all available for further exploration at https://github.com/ohdsi-studies/Covid19Icarius.
These are preliminary findings that are currently in the peer-review process. They should not be relied on to guide clinical practice or health-related behavior and should not be reported in news media as established information.
OHDSI is a multi-stakeholder, interdisciplinary collaborative to bring out the value of health data through large-scale analytics. All solutions are open-source. OHDSI has established an international network of researchers and observational health databases with a central coordinating center housed at Columbia University.
June 5
The OHDSI community has reached an important milestone!
We are excited to announce that Project CHARYBDIS has successfully been tested on multiple databases. We are actively seeking data partners to run CHARYBDIS on their networks. While we are building documentation on this study, any data partners should feel free to reach out to our CHARYBDIS leads (Kristin Kostka, Talita Duarte-Salles and Albert Prats-Uribe) for added assistance.
The protocol and study code is available here: https://github.com/ohdsi-studies/Covid19CharacterizationCharybdis.
As a reminder, Project CHARYBDIS stands for Characterizing Health Associated Risks, and Your Baseline Disease In SARS-COV-2. There are two main objectives for CHARYBDIS: 1) Describe the baseline demographic, clinical characteristics, treatments and outcomes of interest among individuals tested for SARS-CoV-2 and/or diagnosed with COVID-19 overall and stratified by sex, age and specific comorbidities; 2) Describe characteristics and outcomes of hospitalized influenza patients between September 2017 and April 2018 compared to the COVID-19 population.
There are numerous study topics that can be focused within this project, including asthma, cancer, cardiac outcomes, chronic kidney disease, COPD, elderly, end-stage renal disease, gender differences, heart disease, hepatitis C, HIV infection, hypertension, immune disorders, obesity, pediatrics, pregnancy, testing, tuberculosis, type 2 diabetes, and plenty more.
May 28
• We are very excited to announce that the Project CHARYBDIS (Characterizing Health Associated Risks, and Your Baseline Disease In SARS-COV-2) protocol and analysis code have been finalized and uploaded to GitHub. Talita Duarte-Salles, Kristin Kostka and Albert Prats-Uribe are leads for this study, which has two main objectives:
- Describe the baseline demographic, clinical characteristics, treatments and outcomes of interest among individuals tested for SARS-CoV-2 and/or diagnosed with COVID-19 overall and stratified by sex, age and specific comorbidities;
- Describe the characteristics and outcomes of patients diagnosed/tested positive for influenza as well as patients hospitalized with influenza between September 2017 and April 2018 compared to the COVID-19 population.
We have moved this study into the implementation phase and are looking for data partners to run this package. If you have any questions, please reach out to Talita, Kristin or Albert, or check out the FAQ section on our GitHub.
• Patrick Ryan, Daniel Prieto-Alhambra, Talita Duarte-Salles and Ross Williams presented on “OHDSI Community Efforts on COVID-19 Disease Natural History” during Thursday’s Reagan-Udall Foundation for the FDA. Our community representatives were honored to share how OHDSI collaboration and best practices can help study the natural history of COVID-19. If you are interested to learn more about what they discussed, the presentation slides are available here.
May 26
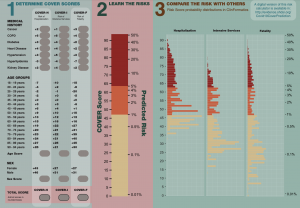 • We are excited to announce that our first Patient-Level Prediction paper has been sent to MedRxiv and is being submitted for peer review. This study “Seek COVER: Development and validation of a personalized risk calculator for COVID-19 outcomes in an international network” is designed to inform individual behavioral choices and help design shielding strategies during de-confinement.
• We are excited to announce that our first Patient-Level Prediction paper has been sent to MedRxiv and is being submitted for peer review. This study “Seek COVER: Development and validation of a personalized risk calculator for COVID-19 outcomes in an international network” is designed to inform individual behavioral choices and help design shielding strategies during de-confinement.
Led by co-first authors Ross Williams and Aniek Markus, the team designed a nine-predictor COVID-19 Estimated Risk (COVER) model that was validated using more than 43,000 COVID patients (following initial development and validation using more than 6.8 million patients with influenza or flu-like symptoms). This model predicts hospitalization, intensive services, and death, and can help provide reassurance for low-risk patients, while shielding high-risk patients, as many start to enter the de-confinement stage of the pandemic.
Peter Rijnbeek is the corresponding author, and both he and Jenna Reps are co-last authors. Overall there are 43 authors involved in the study, once again highlighting the global collaborative nature of the OHDSI community. Congratulations to all who were involved in this work.
• The characterization team plans to post a revised manuscript on MedRxiv with updated data (the original is available here) from its original characterization study on patients hospitalized with COVID-19. The original characterization study was run on four databases, while the most current version has data from eight sources, including a SIDIAP database that includes just under 17,000 COVID patients. This study is the first to systematically summarize medical history in COVID patient populations in the U.S., Europe, and Asia at once.
May 19
• “Shielding the vulnerable: Predicting severe and critical illness in COVID-19: an international multi-database study” is the working title for the first manuscript to come from our prediction studies; this study is analyzing its final datasets before being submitted for publication to both a journal and MedRxiv. Congratulations to Jenna Reps, Ross Williams, Peter Rijnbeek and the entire prediction team on this milestone.
• For those in the MSTeams environment, the lead authors are looking for all co-authorship and COI information by Wednesday, May 20. Check out the posts within the “Study-Prediction” team for more information.
• The phenotype team made tremendous progress at the end of last week to go through Cohort Diagnostics on all cohorts to be used in Project SCYLLA and CHARYBDIS. Phenotypes are being finalized; once finished, packages will be tested at a couple of sites to ensure the viability of the study across our network.
• The original characterization study has added more than 18,000 hospitalized COVID-19 patients from two new datasets, SIDIAP (16,000+) and HM Hospitales (2,000+), both of which come from Spain. These new results are available here. While this study remains under peer review, the team is looking to update the MedRxiv paper with the new results so that the healthcare community can explore this new data. We will update you when a new paper is available.
• Thank you to everybody who provided feedback for the review of the treatments under investigation and the outcomes to evaluation for Project SCYLLA last week. Your input will help us create an even more detailed appendix for both in the protocol.
May 12
• Early results from the external validation of our prediction models have been extremely encouraging. We look forward to further validating the model, including in our newly mapped SIDIAP database. This Spanish database currently includes more than 13,000 COVID-hospitalized patients, which will greatly inform all ongoing COVID-related studies ongoing in the OHDSI community.
• Thank you to all community members who have provided feedback to the treatments and outcomes section of Project SCYLLA. We would like to have all feedback by the end of the week so we can finalize the appendices of the protocol. You can post feedback here.
• Our next characterization study is now being called Project CHARYBDIS (Characterizing Health Associated Risks, and Your Baseline Disease In SARS-COV-2. There are numerous study topics that can be focused within this project, including asthma, cancer, cardiac outcomes, chronic kidney disease, COPD, elderly, end-stage renal disease, gender differences, heart disease, hepatitis C, HIV infection, hypertension, immune disorders, obesity, pediatrics, pregnancy, testing, tuberculosis, type 2 diabetes … and plenty more. Please reach out to project leads Kristin Kostka, Talita Duarte-Salles or Albert Prats-Uribe if you would like to collaborate on this important effort.
• For those less familiar with Greek mythology, SCYLLA and CHARYBDIS were mythical sea monsters that needed to be navigated through in Homer’s epic novel Odyssey. While we certainly have more positive views of our own SCYLLA and CHARYBDIS, we also believe that navigating through both studies will help us get through this modern-day odyssey we are facing. This is our journey, and we believe global collaboration will help us get through our own sea monsters. We hope you will join us in these efforts.
May 7
The Project SCYLLA (SARS-Cov-2 Large-scale Longitudinal Analyses) protocol has been finalized and is available here. SCYLLA’s objectives are to assess the comparative safety and effectiveness of all emerging drug therapies used in COVID-19 treatments …
… administered during hospitalization and prior to intensive services.
… administered during hospitalization after initiating intensive services.
… administered after COVID-19 positive testing and prior to hospitalization.
We believe this set of analyses could provide a critical impact in informing the overall effectiveness of various treatments being used globally against COVID-19. We have databases from 10 nations across Asia, Europe and North America, so we will be able to generate reliable real-world evidence at scale to inform decision-making around this pandemic and are seeking active participation from around the world. We continue to seek more collaborators to join in this important network study; any researchers with data in the OMOP CDM are encouraged to ‘join the journey.’
We have set up a specific thread on the forum seeking community feedback for the treatments and outcomes under investigation; you can find these in Appendix A & B of the protocol.
May 5
The finalized protocol for the “Characterizing and comparing individuals with COVID-19 or influenza” network study is now available in the Characterization channel within the MSTeams environment. As detailed in the protocol, “[t]he primary objective of this study is to describe the baseline demographic and clinical characteristics, as well as treatments and occurrence of outcomes of interest among individuals tested for SARS-CoV-2 or diagnosed with COVID-19 after December 1st 2019, overall and by sex, age and comorbidities. We will also describe the characteristics and outcomes of the population with seasonal influenza infection between September 1st 2016 and April 1st 2018 as a benchmark.”
We are looking for data partners to join in this study. If you are interested, please connect with one of our Principal Investigators (Kristin Kostka, Talita Duarte-Salles, or Albert Prats-Uribe) for more information or assistance in mapping your data to the OMOP Common Data Model.
Thank you to Talita Duarte-Salles for leading the effort to bring in COVID-19 data from Spain. The Hospitals Madrid (HM) dataset currently includes more than 2,000 COVID-19 patients and provides both ongoing and future OHDSI studies COVID-positive patient data from Asia, Europe, and North America.
May 2
A few updates from this week.
• There has been some incredible work already from our phenotype team in advance of our new Characterization and Project Scylla studies. On Tuesday, we began discussion on creating cohorts for these projects, and we have identified a need for more than 150 phenotypes to support our characterization and estimation studies. We are working together to design and evaluate these cohorts. Thanks to everybody in the group for their dedicated work already. For anybody interested in joining this effort, please check out the “Competency-Phenotype development and evaluation” channel.
• Thank you to the community for your feedback to the Project Scylla protocol. Patrick Ryan and Daniel Prieto-Alhambra hope to have an updated version to share next week.
• While our hydroxychloroquine estimation study remains in peer review, the ACE inhibitor/ARBs estimation study is awaiting results from our international set of data partners, including the South Korean HIRA, SIDIAP (Spain), and several U.S. institutions
• The protocol for the IL6/biologics study is under review.
April 29
We are officially one month removed from the end of the #OHDSICOVID19 study-a-thon. Our community continues to build on studies generated from that terrific collaborative event, and we also look towards future studies to aid the COVID-19 response. Still, plenty has happened over the last month, and we wanted to highlight some of it here.
Two studies are currently in the peer-review process, but they have both been posted to MedRxiv. The estimation team completed an important study on the safety of hydroxychloroquine, alone and in combination with azithromycin, the largest study ever on the safety profile of a drug that generated significant attention as a potential COVID-19 therapy. Harlan Krumholz authored a blog post highlighting this study in Forbes.
The characterization team recently submitted the first multi-institution characterization of COVID-19 patients, which reported on the characteristics (demographics, prior conditions and medication use) of more than 6,800 COVID-positive patients from four databases. They were also compared to more than 52,000 patients hospitalized with influenza between 2014-19.
The study-a-thon itself, which was featured here, has many more studies in progress, including several that were discussed during the global closing call. There is an #OHDSICOVID19 YouTube playlist that includes each individual presentation from the global closing call, including studies focused on characterization, estimation and prediction.
Our work is being noticed outside of the OHDSI community. The EMA cited our hydroxychloroquine study in a recent post on the risks involved with HcQ and chloroquine, while both Science Magazine and MedPageToday.com.
Where will we be May 29? As mentioned in the April 28 post, there are exciting initiatives in early development on both characterization studies on various aspects of COVID-19, as well as estimation studies that focus on the safety and anti-viral effectiveness of all emerging therapies for the treatment of COVID-19. Prediction studies are in the process of external validation across our data network, and these could lead to important findings that inform healthcare decision-making around COVID-19. Of course, there is plenty more, so please keep checking this page for regular updates!
April 28
While we continue work generated from the study-a-thon last month, we are excited about some initiatives in development that will help us study COVID-19 and potential treatments more closely.
As discussed last week, Project SCYLLA has been launched to study the safety and anti-viral effectiveness of all emerging therapies for the treatment of COVID-19. Thank you to all community members who provided feedback on the initial protocol; we are working on an updated version, so please check the Study-Estimation-COVID Effectiveness channel in MSTeams for more updates.
The community is also working on characterization studies on various aspects of COVID-19, and an overarching protocol to streamline the work required by data owners to participate in these analyses is also in development. Check out the Study-Characterization channel in MSTeams for more information and updates following the first round of feedback.
Similar to the study-a-thon, the foundation for these studies will be set by the phenotype development team, which began the process of developing approximately 150 cohorts. The first team call to discuss this effort took place this morning on the Competency-Phenotype development and evaluation channel, and collaborators are being sought for this important work.
If you are interested in participating in any of these projects, please head over to those respective channels; if you don’t have MSTeams log-in credentials, please contact Craig Sachson (sachson@ohdsi.org).
April 26
The Observational Health Data Sciences and Informatics (OHDSI) collaboration released a preprint on an international characterization of patients hospitalized with COVID-19 and a comparison with those previously hospitalized with influenza.
Preprints are preliminary reports of work that have not been certified by peer review. They should not be relied on to guide clinical practice or health-related behavior and should not be reported in news media as established information.
The preprint is available here.
The characteristics (demographics, prior conditions and medication use) of more than 6,800 COVID-positive patients from four databases (Columbia, Stanford, the Department of Veterans Affairs and the South Korean Health Insurance Review & Assessment) were reported in this study, and were compared to more than 52,000 patients hospitalized with influenza between 2014-19.
Compared to 52,422 individuals hospitalized with influenza, patients admitted with COVID-19 were more likely male, younger, and, in the US, had fewer comorbidities and lower medication use.
These are preliminary findings that are currently in the peer-review process. They should not be relied on to guide clinical practice or health-related behavior and should not be reported in news media as established information.
OHDSI is a multi-stakeholder, interdisciplinary collaborative to bring out the value of health data through large-scale analytics. All solutions are open-source. OHDSI has established an international network of researchers and observational health databases with a central coordinating center housed at Columbia University.
All updates on OHDSI efforts on COVID-19 will be posted here and within OHDSI community forums, as well as shared on both the community Twitter and LinkedIn feeds.
April 24
Members of the OHDSI community shared this open letter in response to a recent Circulation Research study.
April 23
We are very proud of OHDSI work since last month’s study-a-thon, including several nearly completed manuscripts that we plan to submit for the peer-review process. However, our community also recognizes the critical need for OHDSI to take the next step and leverage its global community, high-level analytics and vast data sources to study which ongoing COVID-19 treatments are actually working.
That brings us to Project SCYLLA.
There is a new channel in MSTeams (Study-Estimation-COVID Effectiveness) where we will collaborate on the design of these studies. Patrick Ryan opened that channel with the post below, and Daniel Prieto-Alhambra followed with a link to the first version of the draft protocol, as well as a request for feedback by 10 am ET on Friday, April 24.
Anybody who was involved with the study-a-thon should still have their MSTeams login and can join the channel; for those who wish to get involved, please reach out to Craig Sachson (sachson@ohdsi.org) for log-in credentials. We are hoping to have community-wide feedback to ensure SCYLLA can generate the highest quality of evidence on therapies around COVID-19.
Below is Patrick’s introductory remarks about Project SCYLLA.
Dear colleagues,
We have been thinking, even before we launched our Study-A-Thon, of the potential to apply our OHDSI tools across the OHDSI network to study the safety and anti-viral effectiveness of all emerging therapies for the treatment of COVID19. Potential therapies against COVID19 are mentioned every day to all of us based on in vitro studies, previous SARS experience and/or experiments, or just clinical experience by colleagues working in different specialties.
Many of us have seen the publication of small observational studies and their impact on clinical practice with great concern. We clearly do not want to contribute to a large amount of research that will be seen in the future as wrong, or even harmful. Hence the reason we hadn’t declared our intention to conduct such studies in the OHDSI community. On the other side, it feels uncomfortable to do nothing if there’s a chance we can positively contribute when so many treatments being actively investigated with limited evidence of any sort about their real-world effects in patients with COVID-19. But much as Odysseus was forced to choose between Scylla and Charybdis on his voyage home, we now need to decide among the lesser of two evils. And following our protagonist’s lead, we also choose Scylla.
We’re announcing here our intention to set forth on another journey, to collaboratively design and implement a population-level effect estimation study to examine the comparative effects of COVID-19 treatments. We’re calling this effort: Project Sc(y)lla: SARS-Cov-2 Large-scale Longitudinal Analyses.
Akin to prior OHDSI LEGEND efforts, our aim is to avoid the pitfalls of cherry-picking specific hypotheses about a particular candidate treatment. Instead, we will create a systematic approach to evaluate all alternative treatments for a large constellation of outcomes of interest in a consistent and reproducible manner. SCYLLA will allow for evidence to accumulate across our network at observational data on COVID-positive patients accrue and additional data partners join the journey to execute our open-source analysis package and share aggregate summary statistics.
Like the Odyssey itself, this will be all about the journey. We recognize the threats and challenges ahead, and we are planning to conduct robust methodological research to test the robustness of our methods while we implement them. These are some of the foreseen difficulties:
- Source-specific outcome/s definition and phenotyping
- Relatively small sample size (by OHDSI standards…)
- Confounding by indication in the midst of continuously emerging therapies
- Prospective data collection
We have prepared a brief slide set (uploaded in the MSTeams page) that will hopefully inspire you to engage with us in the process of designing SCYLLA. We are now writing a first draft study protocol that we will aim to share with you for feedback in the coming few days. Please stay tuned if you are willing to contribute.
Let us start this journey together…
Cheers,
Patrick on behalf of Daniel Prieto Alhambra, Marc Suchard, George Hripcsak, Martijn Schuemie, Martijn, Kristin Kostka, Kristin, Jennifer Lane, Jamie Weaver, Talita Duarte-Salles, Albert Prats-Uribe, Ed Burn and Peter Rijnbeek
April 22
• We are pleased to announce that the manuscript for “Characterization of patients admitted with COVID-19 compared to influenza” has been submitted for peer review. Thank you to Ed Burn for leading this effort, and to our entire characterization team for all the work you put into this important study.
Besides submission for peer review, the manuscript has also been submitted to MedRxiv. Our manuscript on the safety profile of hydroxychloroquine, both alone and in combination with azithromycin, is currently posted on MedRxiv while it undergoes the peer-review process.
There are several standout aspects of this study. It is the first study of COVID-19 hospitalized patients across multiple institutions (Columbia, Stanford, the Veterans Health Administration, and the South Korean HIRA). It is the first paper that provides a summarization of all baseline medical conditions and medication usage of COVID-positive patients, and it is the first to compare hospitalized COVID-positive patients to those hospitalized with influenza (2014-19).
• The prediction models have now all been trained, and we are continuing the process of externally validating them across our data network. If any networks are willing to collaborate in these validations, Jenna Reps and Ross Williams have published a CodeToRun.R that consolidates all prediction protocols into one master run. Documentation related to the four study questions including protocol links can be found here. If you have COVID-19 patients, we hope you will file the protocols with your IRB and contribute to these analyses.
We are encouraged by results from the HIRA database, and we look forward to generating reliable evidence at scale with the assistance of our wide date network. If you have questions, please reach out to Kristin Kostka (kostka@ohdsi.org).
• Our hydroxychloroquine study was profiled in a recent article by Science Magazine: “Antimalarials widely used against COVID-19 heighten risk of cardiac arrest. How can doctors minimize the danger?”
88 Hours: OHDSI’s Signature Moment
The time was meant for highlighting OHDSI capabilities, not testing them.
The hours were meant for sharing global research, not sharing in global research.
The Observational Health Data Sciences and Informatics (OHDSI) community held a COVID-19 global, virtual study-a-thon March 26-29, believing that a network of people who valued both collaboration and open science could make a meaningful impact on the current global pandemic.
How? Nobody was quite sure in the moment, but they were confident they would figure it out.
“We chose an ambitious path and relied on our community and infrastructure to lead the way,” said Patrick Ryan. “In simple terms, efforts within our community over the past 88 months set the foundation for OHDSI’s most important and impactful 88 hours.”
(Click here for the full feature story on the OHDSI COVID-19 study-a-thon)
April 15
The draft for our characterization study is in final review and we hope to submit for peer review by the end of the week. Thank you to Ed Burn for leading this effort and our entire characterization team for the collaborative work on this study.
This was the prioritized study for the characterization team. Once finished, there will be studies that build off this effort, including ones for hospital outcomes and condition occurrences.
The hydroxychloroquine study is currently in the peer-review process, and the preprint was posted on Friday. We are progressing on estimation studies focused on ACE inhibitors and HIV/HepC protease inhibitors. Similarly, the prediction team is advancing on four studies to assist decision-making efforts throughout the global healthcare community; cohorts have been validated for the prediction studies and entered into packages, and the protocols should have greater COVID data to leverage later this week.
We want to thank all of our data partners for their efforts in sharing their data and executing these studies to help generate reliable evidence at scale. The time and expertise that collaborators around the world share daily with OHDSI is a testament to your own dedication in the battle against the COVID-19 pandemic. Together, we are making a difference.
The OHDSI Twitter and LinkedIn feeds continue to replay individual presentations from the final global call; prediction studies have been the focus so far this week, with presentations from Peter Rijnbeek (Monday), Jenna Reps (Tuesday) and Ross Williams (Wednesday) posted so far.
April 10
The Observational Health Data Sciences and Informatics (OHDSI) collaboration released a preprint on preliminary findings from the largest study ever completed on the safety profile of hydroxychloroquine, a drug currently being evaluated as a potential treatment for COVID-19.
Preprints are preliminary reports of work that have not been certified by peer review. They should not be relied on to guide clinical practice or health-related behavior and should not be reported in news media as established information.
The preprint is available here.
The combined short-term use of hydroxychloroquine and azithromycin resulted in nearly 60% higher rate of cardiovascular-related mortality (calHR 2.19; (1.22-3.94)) than the combined use of hydroxychloroquine and amoxicillin. While not as high, there was also an advanced risk for both chest pain/angina (calHR 1.15 (1.05-1.26)) and heart failure (calHR 1.22 (1.02-1.45)) when azithromycin was added to hydroxychloroquine treatment.
These findings were generated from an international database of more than 950,000 users of hydroxychloroquine, including approximately 320,000 who used it in combination with azithromycin.
The short-term effect of hydroxychloroquine as a treatment drug was not found to have an excess risk by itself when compared to sulfasalazine among a large set of patients (950,769 and 306,706, respectively) being treated for rheumatoid arthritis.
Patients from five different countries (Germany, Japan, Spain, the United State, and the United Kingdom) were included in this study, the first to be shared via preprint from a four-day OHDSI COVID-19 study-a-thon, which brought together a global community to design and execute observational studies to generate real-world evidence and help inform the current global pandemic.
These are preliminary findings that are currently in the peer-review process. They should not be relied on to guide clinical practice or health-related behavior and should not be reported in news media as established information.
April 8
The first paper (Safety of hydroxychloroquine with azithromycin: a multi-national study) from our study-a-thon has been submitted for peer review to the New England Journal of Medicine. We are thrilled to have 62 co-authors on the paper, and we believe this could have an immediate and critical impact on clinical practice during this pandemic. Thank you to Jenny Lane and Jamie Weaver for leading the manuscript effort, and to everybody who contributed to this important study.
The characterization team continues to work with data partners across the network on developing source-specific definitions while progressing on a manuscript to describe the baseline characteristics of patients hospitalized with COVID-19. The analysis code is available here. The team is also preparing a characterization analysis to examine treatments and outcomes for these patients, as well as characterizations of patients tested for COVID and those who test COVID-positive. Many in our community are also progressing on more specific characterization studies on different subgroups of COVID patients, with specific efforts to define protocols for pediatrics, cancer, and kidney disease underway. We believe these subgroup analyses will be valuable, and can logically build off of the main characterization analyses that are currently underway as follow-on work.
Various packages for the prediction studies are available here, and they include links to the models and results on the Shiny app. Models for multiple questions have been developed and validated. These studies can impact critical decisions healthcare workers face daily, so thank you to all who have collaborated on this and we look forward to having additional sites perform external validation.
The OHDSI Twitter and LinkedIn feeds will replay individual presentations from the final global call over the next three weeks; over the last two days, presentations on literature review (Jenny Lane) and the OHDSI data network (Kristin Kostka) have been posted.
April 6
The estimation team has prepared a manuscript on the safety profile of hydroxychloroquine that is being submitted for peer review, the pre-print of which would be posted on medrxiv this week; check this page for a link when it becomes available. The team is also progressing on other studies designed during the study-a-thon, including profiles on ACE inhibitors and HIV/HepC protease inhibitors; the ACE team has finalized its protocol, which is posted here.
Our characterization team is working with data partners across the OHDSI network to develop source-specific definitions to identify patients hospitalized with COVID-19 and, once completed, will be compiling aggregate summary statistics that describe the baseline characteristics of those patients. The prediction team has developed prediction models for two questions (‘Amongst Patients Presenting with COVID-19, Influenza, or Associated Symptoms, Who Are Most Likely to be Admitted to the Hospital in the Next 30 Days?’ and ‘Amongst Patients at GP Presenting with Virus or Associated Symptoms with/without Pneumonia Who Are Sent Home, Who Are Most Likely to Require Hospitalization in the Next 30 Days?’), and is seeking support from the OHDSI data partners to externally validate those models in preparation of publication.
The OHDSI Twitter and LinkedIn feeds will replay individual presentations from the final global call over the next two weeks; the introductory presentation by Dani Prieto-Alhambra and Patrick Ryan was posted today and is available here.
April 2
OHDSI has prioritized completing the ‘Baseline Characterization of COVID-19 Patients’ and ‘Safety of Hydroxychloroquine’ studies with a target to summarize results into a draft manuscript by Monday. Additionally, the ACE subteam has been hard at work finalizing its protocol, which is targeted to be posted publicly Friday. The prediction team has been actively working on developing and evaluating models across its 3 questions, with the aim of establishing a simple model that shows sufficient performance and can be used in practice. We will prioritize completing external validation of these PLP models across the OHDSI network after the characterization and HCQ safety studies have been completed.
April 1
OHDSI Collaboration Designs COVID-19 Studies For International Observational Data Network
A four-day global collaboration within the Observational Health Data Sciences and Informatics (OHDSI) community designed and began executing studies on an international set of observational health databases (including insurance claims and electronic health records) to aid decision-making during the current COVID-19 pandemic.
One study is the first large-scale characterization of COVID-19 patients in both the United States and Asia; six databases with COVID-19 patients located in both the U.S. and South Korea already started running data on this project, and other databases are being sought to collaborate in this network study.
The largest study ever conducted on the safety of hydroxychloroquine was designed and executed across an international set of databases. This study of more than 130,000 patients from the USA, England, Germany and South Korea focuses on the overall safety profile of hydroxychloroquine, a drug currently being evaluated as a potential treatment for COVID-19.
The third study designed the first prediction model externally validated on COVID-19 patients to support triage decisions in an effort to ‘flatten the curve’. This model, which determines which patients presenting with symptoms are most likely to require hospitalization, was developed against US data and then tested on South Korean data.
More than 330 people from 30 nations registered to collaborate in this 88-hour virtual study-a-thon, which concluded March 29 with a global presentation from multiple study leads to announce both designs and preliminary findings. Results are currently being evaluated and papers are actively being submitted to journals for peer review.
“It was a humbling effort to lead the OHDSI community in making a meaningful impact during this COVID-19 crisis,” said Daniel Prieto-Alhambra, MD, MSc, PhD, Professor of Pharmaco- and Device Epidemiology at the University of Oxford. “Prioritized questions from governments, health care agencies, and institutions helped direct our efforts, and it was inspiring to see how our community rallied together to make important progress on this research effort.”
Other study designs were presented during the final global update, including safety profiles of other drugs potentially used to treat COVID-19.
OHDSI is a multi-stakeholder, interdisciplinary collaborative to bring out the value of health data through large-scale analytics. All solutions are open-source. OHDSI has established an international network of researchers and observational health databases with a central coordinating center housed at Columbia University.
“I am extremely proud to see what our community accomplished, but we are well aware that this is merely the beginning stage of a long research agenda,” said George Hripcsak, MD, MS, the Vivian Beaumont Allen Professor and Chair of the Columbia Department of Biomedical Informatics. “Our international network is committed to continuing work in this area until this pandemic has ended.”
All updates on OHDSI efforts on COVID-19 will be posted here and within our community forums, and shared on both our Twitter and LinkedIn feeds.
Study-A-Thon Day 4 Updates
Early Call: Video
Global Call: Video
FINAL CALL: Video
More than 340 collaborators.
180 hours.
30 nations over six continents.
17 different teams.
1 global OHDSI community.
This #OHDSICOVID19 study-a-thon was an incredible experience, as we showed the power of global collaboration to set the foundation for our research agenda ahead. The COVID-19 pandemic will not end anytime soon, and our community realizes that much more work is still to come.
The foundation we created over the last four days, however, has given us an amazing head start.
The final update of the OHDSI COVID-19 study-a-thon included presentations around multiple areas of the study-a-thon (these are listed below, with appropriate time slots within the video). Please watch it and see the different characterization, prediction and estimation studies we designed, and some of the early results. Learn about the literature review that informed our study design, and a world-wide data network — including several sources with COVID-19 data — that will execute these studies to help generate real-world evidence.
To our collaborators, we hope you take great pride in what you did over the past four days, and (after a bit of sleep) we hope you find inspiration to continue to collaboratively generate the evidence that promotes better health decisions and better care.
We are OHDSI. That’s what we do.
Again, thank you for your incredible efforts.
PRESENTATIONS WITHIN THE #OHDSICOVID19 WRAP-UP CALL (Full Slidedeck)
Introduction – Daniel Prieto-Alhambra and Patrick Ryan (Slides)
Literature Review – Jennifer Lane (22:00 • Slides)
Data Network In Action – Kristin Kostka (26:10• Slides)
Phenotype Development – Anna Ostropolets (31:38• Slides)
Clinical Characterization of COVID-19 – Ed Burn (42:10 • Slides)
The Journey Through Patient-Level Prediction – Peter Rijnbeek (50:12 • Slides)
Prediction #1: Amongst Patients Presenting with COVID-19, Influenza, or Associated Symptoms, Who Are Most Likely to be Admitted to the Hospital in the Next 30 Days? – Jenna Reps (56:55 • Slides)
Prediction #2: Amongst Patients at GP Presenting with Virus or Associated Symptoms with/without Pneumonia Who Are Sent Home, Who Are Most Likely to Require Hospitalization in the Next 30 Days? – Ross Williams (1:08:42 • Slides)
Prediction #3: Amongst Patients Hospitalized with Pneumonia, Who Are Most Likely To Require Intensive Services or Die? – Aniek Markus (1:15:25 • Slides)
Estimation #1: Hydroxychloroquine – Daniel Prieto-Alhambra (1:23:32 • Slides)
Estimation #2: Safety of HIV/HepC Protease Inhibitors – Albert Prats (1:31:24 • Slides)
Estimation #3: Association of Angiotensin Converting Enzyme (ACE) Inhibitors and Angiotensin II Receptor Blockers (ARB) on COVID Incidence and Complications – Daniel Morales (1:36:58 • Slides)
#OpenData4COVID19 – Seng Chan You (1:45:32 • Slides)
The Journey Ahead – Patrick Ryan (1:50:28 • Slides)
Questions & Answers – Daniel Prieto-Alhambra, Peter Rijnbeek and Patrick Ryan (2:08:15)
Study-A-Thon Day 3 Updates
Early Call: Video
Global Call: Video
Wrap-Up Call: Video
LATE UPDATE
• One night removed from being an empty page, Atlas.ohdsi.org, the OHDSI location for all finalized cohort definitions and analysis packages, now has 113 finalized cohort packages. Prior to being adjudicated and posted, these packages were designed, characterized, evaluated and reviewed by clinical teams. It is important to note (both here and throughout all teams) that quality can not be sacrificed, even in a short time frame. Congratulations to the entire phenotype team for the hours of discussion and work that went into this effort. Patrick Ryan discusses what it takes to define a COVID-19 patient in data around the 6:15 mark of the wrap-up call.
• Huge news from the characterization group, as it has released a study package on Ed Burn’s Github repo. The team took all 12 cohort definitions to characterize hospitalization with COVID-19, as well as (among others) hospitalization with flu during the 2009-10 H1N1 pandemic. This study is being executed across multiple sites in the network. This is a huge milestone, and a show of the importance of collaboration between different teams, as the combined efforts of the phenotyping team and characterization team allowed this study to happen.
• The prediction team finalized a trio of fully-specified prediction questions with completed cohort definition packages. One such question is ‘Among patients who present with flu symptoms, who are the patients who will require admission into the hospital?’ An explanation of that study, as well as others from the prediction team, can be found around the 12:50 mark of the wrap-up call.
• Daniel Prieto-Alhambra takes over around the 22:45 mark of the wrap-up call to discuss the work of the estimation team, with a focus on the study of hydroxychloroquine, which is certainly drawing a great deal of attention right now. This study compares hydroxychloroquine to comparator drugs with a significant dataset, again highlighting the quality of real-world evidence being produced.
Though there is still more work to be done (both in the next 24 hours and beyond), we are so proud and thankful to the hard work of hundreds of collaborators around the world. We believe that this work will inform the healthcare community at a time of critical need, and we are humbled to take this journey with the rest of you!
Early Update
• We have passed the midway point of the weekend, but it’s still ONLY THE BEGINNING of our shared research journey here. The hard work from collaborators around the globe is inspiring, and it’s making a real impact on the different competencies of this virtual study-a-thon. We’re excited to see where we are at the end of the weekend, but we know this will be an incredible foundation for a long journey of research into this global pandemic.
• Daniel Prieto-Alhambra provided updates in both the early and global calls today. The global call focused on planned work for the rest of Saturday. Final phenotypes hope to be agreed upon, and then distributed to the characterization, estimation and prediction teams to use in their respective studies. with The characterization team plans to finalize protocols and complete some analyses. The prediction team will work to train its first model, while the estimation team will be focused on diagnostics, and then launching final analyses.
• We want to commend all the smaller groups within the different teams for their inspired discussions and efforts to move forward on this journey. Collaborators from around the globe find various times to meet on different topics and channels, but there is the same enthusiasm in every call. Thank you to everybody for every ounce of energy you have brought to this research event; it makes a real difference.
Study-A-Thon Day 2 Updates
Early Call: Video
Global Call: Video
Wrap-Up Call: Video
Late Update
• The two main focuses of the Day 2 final call were on data networks and cohort definitions. Patrick Ryan announced that there were 36 data networks over eight different nations that are taking part in the study-a-thon, including seven with some level of COVID-19 data. An early highlight from Day 2 came from the announcement that the South Korean HIRA is making its data available for network studies (see this forum post for more information), but there are also Korean EHR data from two sources. U.S. sources with COVID-19 data include Tufts, Columbia, Stanford and Maine Medical.
• Thanks to Kristin Kostka and Talita Duarte-Salles for their work in helping to prepare these data networks to run the studies currently in preparation, and for joining George Hripcsak and Jose Posada in keeping track of all potential networks with OMOP-standardized data who are ready to contribute to this work.
• There have been 208 cohort definitions developed over the first 30+ hours of the study-a-thon to create populations throughout studies in characterization, estimation and population. As of the update, phenotypes have not been finalized, though the constant conversations have helped direct progress. One of the immediate goals moving forward is finalizing these and sharing them with colleagues across the different specificities.
For more detail and an insightful Q&A session, please check out the Day 2 wrap-up call.
Once again, thank you to everybody who is working so hard these four days to generate critical real-world evidence to inform decision-making during this global pandemic. All the work it took to create an infrastructure to allow this collaboration should be applauded, so we are extra appreciative tonight for the efforts of Lee Evans, Anthony Sena and James Wiggins. Thank you!
Early Update
• The biggest news of the day came during the morning call, as it was announced by Seng Chan You that there was a guide to the new release of COVID-19 data for international collaboration research. Please watch the early call for more details, or read this forum post. If you are interested in participating in the research, please apply here.
At OHDSI, we talk so much about collaboration and open science, and this is such a critical example of those benefits. Thank you to Chan, Rae Woong Park, and everybody in South Korea who worked together to make a national data resource available to enhance the chances of generating meaningful real-world evidence during this global pandemic.
Here are just a few of the team updates so far, but please watch the global call for even more about ongoing progress.
• The estimation team is looking into the effects of several medicines being used in various COVID-19 trials. Protocols are being finalized, with the hopes of analytical packages being distributed within the network later Friday.
• The characterization team continues interesting conversation around how best to characterize a population of hospitalized COVID-19 patients (what symptoms should we be seeking for those who aren’t clearly diagnosed with COVID?). Other challenges have been discussed and addressed, and the team did a great job developing definitions and passing them to the phenotype team.
• The prediction group continues to create definitions for the target population of at-risk patients and the outcomes being studied. Valuable discussions have led to different sensitivity analysis outcome definitions that are being developed. With phenotypes coming (see next note), the group is hard at work developing studies that can provide much-needed insight into healthcare decision-making.
• The phenotype development team continues its work to produce common definitions to share with the afore-mentioned teams for use in upcoming network studies (both this weekend and beyond). By the end of the night, they hope to have all phenotypes necessary for target comparators and outcome cohorts available in a package. It’s motivating to the group to see how its work can impact so many fellow collaborators.
Beyond the update, there was another good Q&A segment at the end of the global call that opened more interesting discussion.
Thank you to all collaborators who continue to power through all the discussions and tasks necessary to go through this particular journey, which will set the foundation of a much longer research journey towards generating real-world evidence that can inform the COVID-19 response. The energy and passion within all the teams is inspiring!
Study-A-Thon Day 1 Updates
Kickoff Call: Video / Slides
Global Call: Video
Wrap-Up Call: Video
PM Update:
It may have only been 16 hours, but there has been tremendous progress already made in our first virtual study-a-thon; the wrap-up call highlighted a lot of the advancements being made throughout the different studies and competencies.
A few notes to come from the wrap-up call.
• Jennifer Lane and Nigel Hughes have led the literature review and protocol development group, and finalized protocols are being developed. Volunteers continue to come forward in this area, which accelerates the process and helps set the foundation for the different study designs being developed.
• Martijn Schuemie has led the process to characterize cohorts in the study package development group. Kristin Kostka and Talita Duarte-Salles are leading the study execution group, which is working with an international set of data partners to prepare for the upcoming network studies. New data sources continue to come forward to share in this critical collaboration!
• The characterization, estimation, prediction and phenotype groups all continue to make strides, and all were discussed in the Day 1 wrap-up call. One focal point was the work done within atlas-covid19.ohdsi.org, which serves as the environment to design concept sets, cohorts and analyses together. A blank page less than 24 hours ago now has more than 100 cohorts in progress, a testament to the efforts of our community.
There was a great Q&A at the end of the wrap-up call which added some insightful discussion, and goals for Day 2 were discussed as well. Thank you to everybody for your hard work in Day 1 on this journey together; we can’t wait to see what is coming next!
AM Update:
• Thank you to everybody who joined us for our kickoff call! It was led by Daniel Prieto-Alhambra (welcome/introduction to the study-a-thon and overview of the COVID-19 pandemic), George Hripcsak (background of OHDSI), Patrick Ryan (what studies will we design/execute during the study-a-thon), and Peter Rijnbeek (logistics of the weekend).
• Following the overall call, there were team kickoff calls on characterization, estimation, prediction and phenotyping. These allowed team leaders to introduce both themselves and OHDSI best practices surrounding each area, and they initiated discussions on the focus of each group throughout the four days. If you are registered for the study-a-thon, the videos of those recordings were uploaded to both the teams and general channels; please watch any or all of them, regardless of what your specific focus is this week.
Thank you to everybody who led these respective calls, including Ed Burn and Ross Williams. We had impressive attendance at all calls, and it didn’t take long for progress to get made. Interesting discussions began immediately within the different channels, and collaborators from around the globe began to take ownership over various responsibilities. A big reason that the study-a-thon could get off to an inspired start was the hard work of the literature review teams, which were discussed in a previous forum post. This early base of knowledge puts the different teams in a strong position to answer important questions on the journey towards study design.
• The global kickoff call was a brief but enthusiastic one, where early progress across the characterization, estimation, prediction and phenotyping group was noted. It was exciting to see so many people — at very different times in their respective days — jump right in. There were also interesting discussions about coordinating virtual communication during this event. OHDSI has experienced impressive success in recent study-a-thons, but this is a new challenge, and some discussion was focused on some practices already being used by the various groups.
• Many people are doing amazing work behind the scenes, and OHDSI would like to highlight a few throughout the weekend. Today, in the crazy rush of getting registrants set for the Microsoft Teams platform and dealing with technical issues, we are thankful for the dedication and hard work by both Henrik John and Mees Mosseveld!
KICKOFF ANNOUNCEMENT
Find the official kick-off press release here
DATA PARTNER VOLUNTEER SURVEY
FORUM UPDATES PRIOR TO THE STUDY-A-THON
March 25: Final thoughts before study-a-thon (ranging from the development of OHDSI to a schedule for the upcoming four days)
March 24 (pm): Update on logistics of study-a-thon
March 24 (am): Reflections on previous OHDSI study-a-thons, and what we learned to strengthen our current effort
March 23: Update on new vocabulary, reminders on data survey, registration
March 22: Update on registration closing, data survey
March 20: Update
March 17: Update
March 7: Original announcement + a status update one week later (midway down thread)


