- Who We Are
- Updates & News
- Standards
- Software Tools
- Network Studies
- Community Forums
- Education
- New To OHDSI?
- Community Calls
- Past Events
- 2024 Europe Symposium
- 2024 Thailand Tutorial
- 2024 Japan Tutorial
- 2024 DevCon
- 2024 Phenotype Phebruary
- 2023 Global Symposium
- 2023 APAC Symposium
- 2023 Europe Symposium
- 2023 SOS Challenge
- 2023 DevCon
- 2023 Phenotype Phebruary
- 2022 Global Symposium
- 2021 Global Symposium
- 2020 Global Symposium
- ALL Past OHDSI Events
- Workgroups
- 2023 ‘Our Journey’ Annual Report
- This Week In OHDSI
- Support & Sponsorship
- CBER Best Seminars
- 2024 Europe Symposium
- 2024 Global Symposium
- Github
- YouTube
- Newsletters
OHDSI Community Calls
Everybody is invited to the weekly OHDSI community call, which takes place each Tuesday at 11 am ET. These calls are meant to inform and engage our community through a variety of call formats, including community presentations, workgroup updates, breakout sessions, publication announcements, newcomer-focused sessions, and more. The upcoming schedule is available to the right.
Videos, slides and weekly updates from this year’s calls are available below. Presentations from the 2023, 2022 and 2021 community calls are also available.
Weekly Recordings & Updates
The July 23 community call featured the second of four summer sessions on Building Up The OHDSI Evidence Network. Clair Blacketer, Paul Nagy, and Ben Martin led a session focused on Evidence Network details, IRB language and strategies, a live demo focused on how to join the Evidence Network, and some Q&A throughout following questions from other community members.
Community Updates
• Congratulations to the team of Tom Seinen, Jan Kors, Erik van Mulligen, and Peter Rijnbeek on the publication of Annotation-preserving machine translation of English corpora to validate Dutch clinical concept extraction tools in JAMIA.
• The agenda for the main conference day of the 2024 OHDSI Global Symposium is now available. The August 13 community call will focus on both the main conference day and a look at the individual tutorials.
• The Eye Care & Vision Research workgroup is leading a Semaglutide NAION network study based on last week’s OHDSI Evidence Network presentation. Cindy Cai and Michelle Hribar discussed this study and the value of network collaboration; please use this link to join the Eye Care & Vision Workgroup.
• The fifth European OHDSI Symposium, titled “Scaling up Reliable Evidence Across Europe,” was held June 1-3 and brought together data partners, regulators, and researchers to collaborate and share results and ideas about the use of the OMOP-CDM in Europe. All materials from the session, including talks focused on the Selection of European Initiatives Using the OMOP CDM and Large Scale Evidence Generation in EHDEN and DARWIN EU®, as well as the collaborator showcase, can be found all on the event homepage.
OHDSI Social Showcase
• Research from the 2024 Europe Symposium Collaborator Showcase is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — External Validation of the Revised Cardiac Risk Index (RCRI) Clinical Prediction Model in Observational Health Care Databases (Alexander Saelmans)
Tuesday — Conversion of the Papageorgiou General Hospital EHR to the OMOP Common Data Model (Papapostolou Grigoris)
Wednesday — Automated OMOP-CDM pipeline for the new EBMT Registry (Shirah Cashriel)
Thursday — Incorporating Temporal Information from EHR Data in Clinical Prediction Modelling (Estelle Lampel)
Friday — Defining international approaches for the detection of emergent metastasis and the classification of location of metastasis from hospital EHR (Stelios Theophanous)
2024 OHDSI Global Symposium
• Registration is open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
Job Openings
• Ross Williams announced an opening for a Health Data Scientist at Erasmus MC. The candidate will be responsible for the creation and execution of study software to support the work of epidemiologists. This will include designing, developing, documenting, and maintaining R code that will be executed against health data that is standardized to the OMOP Common Data Model. More information and an application link are available here.
• Ajit Londhe announced an opening for a Senior Associate Director, Real World Evidence and Analytics at Boehringer Ingelheim. The candidate for this remote position will have the opportunity to generate real world evidence (RWE) to support in-line and pipeline products, provide statistical advice on the analysis of real world data (RWD) to various internal and external stakeholders, contribute to the RWD acquisition strategy and tool evaluation, and participate in the development and presentation of RWE trainings. More information and an application link is available here.
• Kevin Haynes shared an opening at CVS Health for a Lead Director, RWE Distributed Research. This position will lead a team of Analytic professionals responsible for transformation of healthcare insurance and pharmacy data into distributed analytic models, including OMOP, used in safety surveillance and collaborative research (SS&C). More information and an application link is available here.
• Aki Nishimura announced that Johns Hopkins University is seeking postdoctoral fellows. The fellows would work on methodological research in pharmaco-epidemiology to address medication and device utilization, effectiveness, and safety relevant to health, lung, and blood diseases. More information and application details are available here. If you are interested in having Aki Nishimura as a co-advisor, please include him on your application email at aki.nishimura@jhu.edu.
Slides
Building The OHDSI Evidence Network | Community Updates
Video Presentation
 The July 16 (11 am ET) community call focused on the HowOften Initiative, including early findings following the 2023 OHDSI Global Symposium workshop, different projects using HowOften, including one related to clinicaltrials.gov data, and future steps. Below are the leads for the different talks; slides and videos from the session are located below.
The July 16 (11 am ET) community call focused on the HowOften Initiative, including early findings following the 2023 OHDSI Global Symposium workshop, different projects using HowOften, including one related to clinicaltrials.gov data, and future steps. Below are the leads for the different talks; slides and videos from the session are located below.
• Hsin Yi “Cindy” Chen (MD-PhD Student, Columbia University Department of Biomedical Informatics)
• Azza Shoaibi (Director, Observational Health Data Analytics, Janssen Research and Development)
• Elise Ruan (Clinical Informatics Fellow, NewYork-Presbyterian Hospital/Columbia University)
• George Hripcsak (Professor of Biomedical Informatics, Columbia University)
Community Updates
• Congratulations to the team of Jason Patterson and Nicholas Tatonetti on the publication of KG-LIME: predicting individualized risk of adverse drug events for multiple sclerosis disease-modifying therapy in JAMIA.
• Congratulations to the team of Yong Shang, Yu Tian, Kewei Lyu, Tianshu Zhou, Ping Zhang, Jianghua Chen, and Jingsong Li on the publication of Electronic Health Record–Oriented Knowledge Graph System for Collaborative Clinical Decision Support Using Multicenter Fragmented Medical Data: Design and Application Study in the Journal of Medical Internet Research.
• The Eye Care & Vision Research workgroup is leading a Semaglutide NAION network study based on last week’s OHDSI Evidence Network presentation. Cindy Cai and Michelle Hribar discussed this study and the value of network collaboration; please use this link to join the Eye Care & Vision Workgroup.
• The next CBER BEST Seminar will be held Wed., July 17, at 11 am ET and will be led by Yonas Ghebremichael-Weldeselassie, Lecturer of Statistics at School of Mathematics and Statistics, The Open University, UK. The focus of this talk will be A modified self-controlled case series method for event-dependent exposures and high event-related mortality, with application to COVID-19 vaccine safety. You can access the session using this link.
• The fifth European OHDSI Symposium, titled “Scaling up Reliable Evidence Across Europe,” was held June 1-3 and brought together data partners, regulators, and researchers to collaborate and share results and ideas about the use of the OMOP-CDM in Europe. All materials from the session, including talks focused on the Selection of European Initiatives Using the OMOP CDM and Large Scale Evidence Generation in EHDEN and DARWIN EU®, as well as the collaborator showcase, can be found all on the event homepage.
OHDSI Social Showcase
• Research from the 2024 Europe Symposium Collaborator Showcase is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Advancing Certification and Evaluation of Medical Device Software in the EU using OMOP (Frédéric Jung)
Tuesday — SNOMED overhaul and its impact on ETL and phenotyping (Masha Khitrun)
Wednesday — OMOPification of real world cancer data to enable privacy-preserving analytics for cancer research (Prabash Galgane Banduge)
Thursday — The association between comorbid depression and insulin initiation in type 2 diabetes A cohort OHDSI study (Christianus Heru Setiawan)
Friday — Empowering research with seamless data flow and research-ready, anonymised data in OMOP CDM: Learnings from the design of WAYFIND-R, a global precision oncology registry and research platform (Tom Stone)
2024 OHDSI Global Symposium
• Registration is open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
Job Openings
• Ross Williams announced an opening for a Health Data Scientist at Erasmus MC. The candidate will be responsible for the creation and execution of study software to support the work of epidemiologists. This will include designing, developing, documenting, and maintaining R code that will be executed against health data that is standardized to the OMOP Common Data Model. More information and an application link are available here.
• Ajit Londhe announced an opening for a Senior Associate Director, Real World Evidence and Analytics at Boehringer Ingelheim. The candidate for this remote position will have the opportunity to generate real world evidence (RWE) to support in-line and pipeline products, provide statistical advice on the analysis of real world data (RWD) to various internal and external stakeholders, contribute to the RWD acquisition strategy and tool evaluation, and participate in the development and presentation of RWE trainings. More information and an application link is available here.
• Kevin Haynes shared an opening at CVS Health for a Lead Director, RWE Distributed Research. This position will lead a team of Analytic professionals responsible for transformation of healthcare insurance and pharmacy data into distributed analytic models, including OMOP, used in safety surveillance and collaborative research (SS&C). More information and an application link is available here.
• Aki Nishimura announced that Johns Hopkins University is seeking postdoctoral fellows. The fellows would work on methodological research in pharmaco-epidemiology to address medication and device utilization, effectiveness, and safety relevant to health, lung, and blood diseases. More information and application details are available here. If you are interested in having Aki Nishimura as a co-advisor, please include him on your application email at aki.nishimura@jhu.edu.
Slides
HowOften: Background | Trends in Hospitalization Among Patients with Cardiovascular, Immunological, and neurological Illnesses | Does Indication Matter? | Standardization of Clinical Trials Data for Large-Scale Incidence Calculations of Adverse Drug Events | Future Steps
Video Presentation
00:40 – Background (Chen)
04:22 – Trends in Hospitalization Among Patients with Cardiovascular, Immunological, and neurological Illnesses (Shoaibi)
15:58 – Does Indication Matter? (Chen)
25:56 – Standardization of Clinical Trials Data for Large-Scale Incidence Calculations of Adverse Drug Events (Ruan)
40:28 – Future Steps (Hripcsak)
July 9: Building the OHDSI Evidence Network, Session 1 - Introduction, Value to the Community (Ryan)
 The July 9 community call featured the first of four summer sessions on Building Up The OHDSI Evidence Network. Patrick Ryan led a session that introduced the OHDSI Evidence Network and how it can benefit the community mission, the challenges around getting OMOP data sources to engage in network studies, a recently published study that could be tested through the Evidence Network, and more.
The July 9 community call featured the first of four summer sessions on Building Up The OHDSI Evidence Network. Patrick Ryan led a session that introduced the OHDSI Evidence Network and how it can benefit the community mission, the challenges around getting OMOP data sources to engage in network studies, a recently published study that could be tested through the Evidence Network, and more.
Community Updates
• Congratulations to the team of Jason Patterson and Nicholas Tatonetti on the publication of KG-LIME: predicting individualized risk of adverse drug events for multiple sclerosis disease-modifying therapy in JAMIA.
• Congratulations to the team of Yong Shang, Yu Tian, Kewei Lyu, Tianshu Zhou, Ping Zhang, Jianghua Chen, and Jingsong Li on the publication of Electronic Health Record–Oriented Knowledge Graph System for Collaborative Clinical Decision Support Using Multicenter Fragmented Medical Data: Design and Application Study in the Journal of Medical Internet Research.
• The fifth European OHDSI Symposium, titled “Scaling up Reliable Evidence Across Europe,” was held June 1-3 and brought together data partners, regulators, and researchers to collaborate and share results and ideas about the use of the OMOP-CDM in Europe. All materials from the session, including talks focused on the Selection of European Initiatives Using the OMOP CDM and Large Scale Evidence Generation in EHDEN and DARWIN EU®, as well as the collaborator showcase, can be found all on the event homepage.
• The next CBER BEST Seminar will be held Wed., July 17, at 11 am ET and will be led by Yonas Ghebremichael-Weldeselassie, Lecturer of Statistics at School of Mathematics and Statistics, The Open University, UK. The focus of this talk will be A modified self-controlled case series method for event-dependent exposures and high event-related mortality, with application to COVID-19 vaccine safety. You can access the session using this link.
• Sarah Seager is the Senior Director, Analytics & AI at IQVIA. She is an experienced technical senior leader who develops and leads analytical teams in the world of data science and advanced analytics. In the latest edition of the Collaborator Spotlight, Sarah shares her thoughts on the current path of data management and analytics, why OMOP is an ideal common data model, recent advances around the European community, and how data is similar to another one of Sarah’s passions, art.
• To carry out the OHDSI mission, we need an active and willing global network of data partners, and we need the ability to quickly identify those that might be the right fit for a specific clinical research question. Last year we piloted this effort through the Save our Sisyphus challenge and are now ready to move forward based on our learnings. The OHDSI Evidence Network workgroup is excited to initiate a network study that will describe the OHDSI Network in a publication, and will also create an open public resource designed to facilitate evidence generation faster and better than ever by building on methodologies developed by thought leaders around the world. You can access the protocol here, and you can learn more about this effort from a recent OHDSI Evidence Network update. Come join us on this exciting journey!
OHDSI Social Showcase
• Research from the 2024 Europe Symposium Collaborator Showcase is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Additional technical data protection measures to improve the security of an OMOP/OHDSI infrastructure (Francesco Pozzoni)
Tuesday — Forecasting the prescription rates of antibiotics in the UK between 2013 to 2023 incorporating the impact of COVID-19 (Yuchen Guo)
Wednesday — Challenges in harmonising data across multiple biobanks (Karyn Mégy)
Thursday — Predicting long term cancer survival for Health Technology Assessment: A multinational cohort study across Europe (Jeremy Dietz)
Friday — CDM Onboarding R package for data quality assessment (Anne van Winzum)
2024 OHDSI Global Symposium
• Registration is open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
Job Openings
• Ross Williams announced an opening for a Health Data Scientist at Erasmus MC. The candidate will be responsible for the creation and execution of study software to support the work of epidemiologists. This will include designing, developing, documenting, and maintaining R code that will be executed against health data that is standardized to the OMOP Common Data Model. More information and an application link are available here.
• Ajit Londhe announced an opening for a Senior Associate Director, Real World Evidence and Analytics at Boehringer Ingelheim. The candidate for this remote position will have the opportunity to generate real world evidence (RWE) to support in-line and pipeline products, provide statistical advice on the analysis of real world data (RWD) to various internal and external stakeholders, contribute to the RWD acquisition strategy and tool evaluation, and participate in the development and presentation of RWE trainings. More information and an application link is available here.
• Kevin Haynes shared an opening at CVS Health for a Lead Director, RWE Distributed Research. This position will lead a team of Analytic professionals responsible for transformation of healthcare insurance and pharmacy data into distributed analytic models, including OMOP, used in safety surveillance and collaborative research (SS&C). More information and an application link is available here.
• Aki Nishimura announced that Johns Hopkins University is seeking postdoctoral fellows. The fellows would work on methodological research in pharmaco-epidemiology to address medication and device utilization, effectiveness, and safety relevant to health, lung, and blood diseases. More information and application details are available here. If you are interested in having Aki Nishimura as a co-advisor, please include him on your application email at aki.nishimura@jhu.edu.
Slides
Building The OHDSI Evidence Network | Community Updates
Video Presentation
During our annual “Meet New Members of the OHDSI Community” session, some of our newest collaborators had the opportunity to introduce themselves, share their professional journey and discuss how they hope to impact the OHDSI community.
Community Updates
• Congratulations to the team of Fateme Nateghi Haredasht, Sajjad Fouladvand, Steven Tate, Min Min Chan, Joannas Jie Lin Yeow, Kira Griffiths, Ivan Lopez, Jeremiah W. Bertz, Adam S. Miner, Tina Hernandez-Boussard, Chwen-Yuen Angie Chen, Huiqiong Deng, Keith Humphreys, Anna Lembke, L. Alexander Vance, and Jonathan H. Chen on the publication of Predictability of buprenorphine-naloxone treatment retention: A multi-site analysis combining electronic health records and machine learning in Addiction.
• Congratulations to the team of Gyunam Park, Yaejin Lee, and Minsu Cho on the publication of Enhancing healthcare process analysis through object-centric process mining: Transforming OMOP common data models into object-centric event logs in the Journal of Biomedical Informatics.
• The latest edition of the OHDSI newsletter is now available. It includes information on the OHDSI Evidence Network, LLMs in the evidence generation process, the latest collaborator spotlight with Sarah Seager, recent publications and presentations, community updates and more.
• To carry out the OHDSI mission, we need an active and willing global network of data partners, and we need the ability to quickly identify those that might be the right fit for a specific clinical research question. Last year we piloted this effort through the Save our Sisyphus challenge and are now ready to move forward based on our learnings. The OHDSI Evidence Network workgroup is excited to initiate a network study that will describe the OHDSI Network in a publication, and will also create an open public resource designed to facilitate evidence generation faster and better than ever by building on methodologies developed by thought leaders around the world. You can access the protocol here, and you can learn more about this effort from a recent OHDSI Evidence Network update. Come join us on this exciting journey!
• The workshop on “AI for Reliable and Equitable Real-World Evidence Generation in Medicine” at the International Conference on Artificial Intelligence in Medicine (AIME) will take place on July 9, 2024 at Salt Lake City, Utah. The workshop focuses on advancing the understanding and exploring the transformative role of artificial intelligence (AI) in analyzing real-world data (RWD) for real-world evidence (RWE) generation. Speakers and panelists from informatics, biostatistics, computer science, and related fields across academic institutions, federal agencies, and healthcare providers will be joining us for the workshop.
OHDSI Social Showcase
• Research from the 2024 Europe Symposium Collaborator Showcase is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday —Transforming lung cancer EHR data into the OMOP CDM: A case study of Non Small Cell Lung Cancer (Evangelos Chandakas (Handakas))
Tuesday — Four Complexities when mapping NCRAS to the OMOP CDM (Laura Kerr)
Wednesday — Piloting the Transformation of Multiple Sclerosis Real-World Data to the OMOP CDM: Lessons Learned (Tina Parciak)
Thursday — Towards all-Island sharing of Irish lymphoid blood cancers data: landscape and gap analysis (Kluivert Boakye Duah)
Friday — Exploring Drug Utilization Patterns in Osteoporosis Therapy (Balqis Istiqomah Gusbela)
2024 OHDSI Global Symposium
• Registration is open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
Job Openings
• Ajit Londhe announced an opening for a Senior Associate Director, Real World Evidence and Analytics at Boehringer Ingelheim. The candidate for this remote position will have the opportunity to generate real world evidence (RWE) to support in-line and pipeline products, provide statistical advice on the analysis of real world data (RWD) to various internal and external stakeholders, contribute to the RWD acquisition strategy and tool evaluation, and participate in the development and presentation of RWE trainings. More information and an application link is available here.
• Kevin Haynes shared an opening at CVS Health for a Lead Director, RWE Distributed Research. This position will lead a team of Analytic professionals responsible for transformation of healthcare insurance and pharmacy data into distributed analytic models, including OMOP, used in safety surveillance and collaborative research (SS&C). More information and an application link is available here.
• Aki Nishimura announced that Johns Hopkins University is seeking postdoctoral fellows. The fellows would work on methodological research in pharmaco-epidemiology to address medication and device utilization, effectiveness, and safety relevant to health, lung, and blood diseases. More information and application details are available here. If you are interested in having Aki Nishimura as a co-advisor, please include him on your application email at aki.nishimura@jhu.edu.
Slides
Video Presentation
Meet The Newcomers
 The June 25 community call featured our quarterly ‘Recent Publications’ theme. We were happy to welcome four lead authors of recent studies that highlighted either OMOP or OHDSI tools/practices.
The June 25 community call featured our quarterly ‘Recent Publications’ theme. We were happy to welcome four lead authors of recent studies that highlighted either OMOP or OHDSI tools/practices.
• Nhung Trinh, Researcher, University of Oslo
Effectiveness of COVID-19 vaccines to prevent long COVID: data from Norway (The Lancet Respiratory Medicine)
• Theresa Burkard, Postdoctoral Data Scientist, University of Oxford
Calculating daily dose in the Observational Medical Outcomes Partnership Common Data Model (Pharmacoepidemiology & Drug Safety)
• Egill Fridgeirsson, Scientific Researcher, Erasmus MC
Comparing penalization methods for linear models on large observational health data (JAMIA)
• Cindy Cai, Assistant Professor of Ophthalmology, Johns Hopkins University
Similar Risk of Kidney Failure among Patients with Blinding Diseases Who Receive Ranibizumab, Aflibercept, and Bevacizumab: An Observational Health Data Sciences and Informatics Network Study (Ophthalmology Retina)
Community Updates
• Congratulations to the team of Nora Tabea Sibert, Johannes Soff, Sebastiano La Ferla, Maria Quaranta, Andreas Kremer, Christoph Kowalski on the publication of Transforming a Large-Scale Prostate Cancer Outcomes Dataset to the OMOP Common Data Model—Experiences from a Scientific Data Holder’s Perspective in Cancers.
• Congratulations to the team of Egill Fridgeirsson, Ross Williams, Peter Rijnbeek, Marc Suchard, and Jenna Reps on the publication of Comparing penalization methods for linear models on large observational health data in JAMIA.
• Congratulations to the team of Katja Hoffmann, Igor Nesterow, Yuan Peng, Elisa Henke, Daniela Barnett, Cigdem Klengel, Mirko Gruhl, Martin Bartos, Frank Nüßler, Richard Gebler, Sophia Grummt, Anne Seim, Franziska Bathelt, Ines Reinecke, Markus Wolfien, Jens Weidner, and Martin Sedlmayr on the recent publication of Streamlining intersectoral provision of real-world health data: a service platform for improved clinical research and patient care in Frontiers of Medicine.
• Congratulations to the team of Nicolas Alexander Schulz, Jasmin Carus, Alexander Johannes Wiederhold, Ole Johanns, Frederik Peters, Natalie Rath, Katharina Rausch, Bernd Holleczek, Alexander Katalinic, the AI-CARE Working Group & Christopher Gundler on the recent publication of Learning debiased graph representations from the OMOP common data model for synthetic data generation in BMC Medical Research Methodology.
• Thank you to everybody who shared brief reports for the 2024 Global Symposium. We had more than 140 submissions this year, including more than 20 software demos. The review process will begin this week when the scientific review committee meets this Thursday at 11 am ET.
• To carry out the OHDSI mission, we need an active and willing global network of data partners, and we need the ability to quickly identify those that might be the right fit for a specific clinical research question. Last year we piloted this effort through the Save our Sisyphus challenge and are now ready to move forward based on our learnings. The OHDSI Evidence Network workgroup is excited to initiate a network study that will describe the OHDSI Network in a publication, and will also create an open public resource designed to facilitate evidence generation faster and better than ever by building on methodologies developed by thought leaders around the world. You can access the protocol here, and you can learn more about this effort from a recent OHDSI Evidence Network update. Come join us on this exciting journey!
• The next edition of the CBER BEST Seminar Series will be held Wednesday, June 26, at 11 am ET. Jenna Wong, Assistant Professor in the Department of Population Medicine at Harvard Medical School and Harvard Pilgrim Health Care Institute, will lead a session on Applying Machine Learning in Distributed Networks to Support Activities for Post-Market Surveillance of Medical Products: Opportunities, Challenges, and Considerations. The full schedule for the CBER BEST Seminar Series, including past presentations, is available here.
• The July 2 community call will be our annual “Newcomer Introductions” session. We invite any of our newer members of the OHDSI community to join the call, introduce themselves and share about how they hope to collaborate with the community. To ensure the opportunity to take part in this session, please fill out this brief form.
• The workshop on “AI for Reliable and Equitable Real-World Evidence Generation in Medicine” at the International Conference on Artificial Intelligence in Medicine (AIME) will take place on July 9, 2024 at Salt Lake City, Utah. The workshop focuses on advancing the understanding and exploring the transformative role of artificial intelligence (AI) in analyzing real-world data (RWD) for real-world evidence (RWE) generation. Speakers and panelists from informatics, biostatistics, computer science, and related fields across academic institutions, federal agencies, and healthcare providers will be joining us for the workshop.
OHDSI Social Showcase
• Research from the 2024 Europe Symposium Collaborator Showcase is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Expanding the OUH Clinical Data Warehouse data delivery infrastructure with OMOP CDM and OHDSI tools (Olivier Bouissou)
Tuesday — Trade-offs in the design of explainable prediction models for health care (Aniek Markus)
Wednesday — BC Platforms’ Systematic Approach to OMOP Oncology Federated Query and Analytical Solutions in ONCOVALUE consortium (Mai Nguyen)
Thursday — Create a matched cohort to provide context for your large-scale characterization (Marta Alcalde-Herraiz)
Friday — Advancements in Automated OMOP Concept Code Selection: Leveraging GPT-4 for Efficient and Accurate Mapping of Drug description and ATC to RxNorm (Extension) (Vita De Vos)
2024 OHDSI Global Symposium
• Registration is open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
Job Openings
• Daniel Prieto-Alhambra announced an opening for a Postdoctoral Researcher in Real World Evidence to join the Pharmaco- and Device epidemiology research group at the Botnar Research Centre, NDORMS, University of Oxford. This person will be leading or co-leading real world evidence studies, analysing real world health data mapped to the OMOP common data model and write study reports and scientific manuscripts. More details and an application link are available here; the application deadline is 12 pm on July 1, 2024.
• Aki Nishimura announced that Johns Hopkins University is seeking postdoctoral fellows. The fellows would work on methodological research in pharmaco-epidemiology to address medication and device utilization, effectiveness, and safety relevant to health, lung, and blood diseases. More information and application details are available here. If you are interested in having Aki Nishimura as a co-advisor, please include him on your application email at aki.nishimura@jhu.edu.
Slides
Huang | Burkard | Fridgeirsson | Cai | Community Updates
Video Presentation
Effectiveness of COVID-19 vaccines to prevent long COVID: data from Norway (Nhung Trinh)
Calculating daily dose in the Observational Medical Outcomes Partnership Common Data Model (Theresa Burkard)
Comparing penalization methods for linear models on large observational health data (Egill Fridgeirsson)
Similar Risk of Kidney Failure among Patients with Blinding Diseases Who Receive Ranibizumab, Aflibercept, and Bevacizumab: An Observational Health Data Sciences and Informatics Network Study (Cindy Cai)
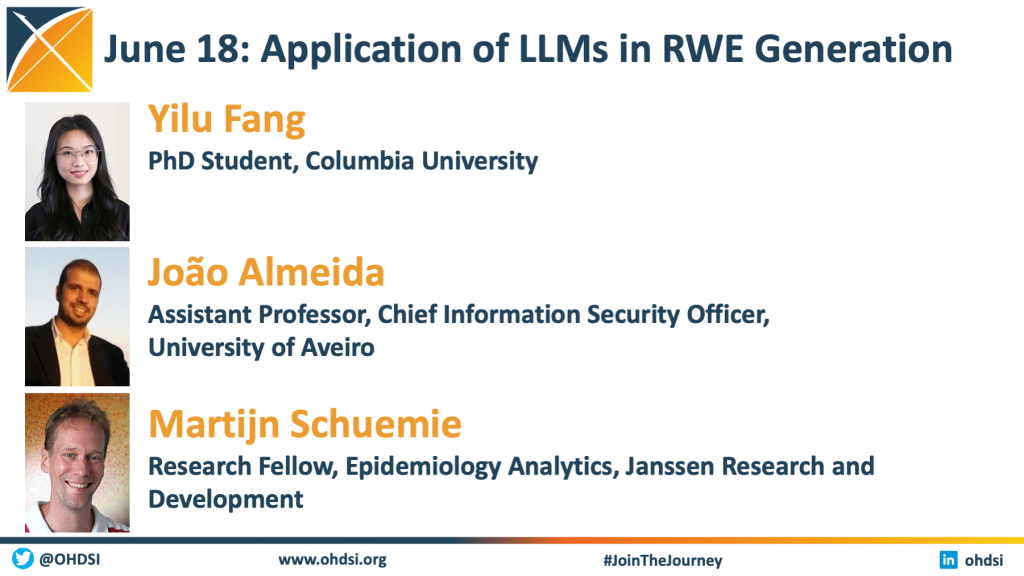 The June 18 community call focused on the Application of Large Language Models in the Evidence Generation Process. We welcomed three members of the community to present recent research in this area.
The June 18 community call focused on the Application of Large Language Models in the Evidence Generation Process. We welcomed three members of the community to present recent research in this area.
Knowledge-guided Generative AI For Automated Taxonomy Learning From Drug Labels – Yilu Fang • PhD Student, Columbia University
A Chatbot to Streamline Biomedical Data Discovery and Analysis – João Almeida • Chief Information Security Officer, University of Aveiro
Generative AI for real-world evidence – Martijn Schuemie • Research Fellow, Epidemiology Analytics, Janssen Research and Development
Community Updates
• All collaborator showcase submissions for the 2024 Global Symposium are due by 8 pm ET on Friday, June 21. Please use this link to submit your brief report!
• To carry out the OHDSI mission, we need an active and willing global network of data partners, and we need the ability to quickly identify those that might be the right fit for a specific clinical research question. Last year we piloted this effort through the Save our Sisyphus challenge and are now ready to move forward based on our learnings. The OHDSI Evidence Network workgroup is excited to initiate a network study that will describe the OHDSI Network in a publication, and will also create an open public resource designed to facilitate evidence generation faster and better than ever by building on methodologies developed by thought leaders around the world.
You can access the protocol here, and you can learn more about this effort from a recent OHDSI Evidence Network update. Come join us on this exciting journey!
• The next edition of the CBER BEST Seminar Series will be held Wednesday, June 26, at 11 am ET. Jenna Wong, Assistant Professor in the Department of Population Medicine at Harvard Medical School and Harvard Pilgrim Health Care Institute, will lead a session on Applying Machine Learning in Distributed Networks to Support Activities for Post-Market Surveillance of Medical Products: Opportunities, Challenges, and Considerations. The full schedule for the CBER BEST Seminar Series, including past presentations, is available here.
• The July 2 community call will be our annual “Newcomer Introductions” session. We invite any of our newer members of the OHDSI community to join the call, introduce themselves and share about how they hope to collaborate with the community. To ensure the opportunity to take part in this session, please fill out this brief form.
2024 OHDSI Global Symposium
• Registration is open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
• Collaborator Showcase submissions are now being accepted, and all details about the event are available here. Submissions are due by Friday, June 21, at 8 pm ET. Notifications of acceptance will be sent out by Tuesday, Aug. 20.
Job Openings
• Daniel Prieto-Alhambra announced an opening for a Postdoctoral Researcher in Real World Evidence to join the Pharmaco- and Device epidemiology research group at the Botnar Research Centre, NDORMS, University of Oxford. This person will be leading or co-leading real world evidence studies, analysing real world health data mapped to the OMOP common data model and write study reports and scientific manuscripts. More details and an application link are available here; the application deadline is 12 pm on July 1, 2024.
• Aki Nishimura announced that Johns Hopkins University is seeking postdoctoral fellows. The fellows would work on methodological research in pharmaco-epidemiology to address medication and device utilization, effectiveness, and safety relevant to health, lung, and blood diseases. More information and application details are available here. If you are interested in having Aki Nishimura as a co-advisor, please include him on your application email at aki.nishimura@jhu.edu.
Slides
Feng | Almeida | Schuemie | Community Updates
Video Presentation
Knowledge-guided Generative AI For Automated Taxonomy Learning From Drug Labels (Yilu Fang)
A Chatbot to Streamline Biomedical Data Discovery and Analysis (João Almeida)
Generative AI for real-world evidence (Martijn Schuemie)
 The June 11 community call provided a review of the 2024 OHDSI Europe Symposium. Following an introduction about the event, we heard about the training activities (with a focus on ETL), software demos, and an example of progress being made among the National Nodes (Belgium).
The June 11 community call provided a review of the 2024 OHDSI Europe Symposium. Following an introduction about the event, we heard about the training activities (with a focus on ETL), software demos, and an example of progress being made among the National Nodes (Belgium).
We were excited to welcome the following to lead this session:
– Peter Reinbeek • Professor of Medical Informatics and Chair of the Department of Medical Informatics, Erasmus MC
– Maxim Moinat • Scientific Researcher, Erasmus MC
– Cesar Barboza Gutierrez • R Software Developer, Erasmus MC
– Liesbet Peeters • Assistant Professor of Biomedical Data Sciences, UHasselt
Clair Blacketer also provided an update on a new protocol and network study associated with the OHDSI Evidence Network, and shared how databases can join this growing research asset for the community.
Both video presentations are available below.
Community Updates
• All collaborator showcase submissions for the 2024 Global Symposium are due by 8 pm ET on Friday, June 21. Please use this link to submit your brief report!
• To carry out the OHDSI mission, we need an active and willing global network of data partners, and we need the ability to quickly identify those that might be the right fit for a specific clinical research question. Last year we piloted this effort through the Save our Sisyphus challenge and are now ready to move forward based on our learnings. The OHDSI Evidence Network workgroup is excited to initiate a network study that will describe the OHDSI Network in a publication, and will also create an open public resource designed to facilitate evidence generation faster and better than ever by building on methodologies developed by thought leaders around the world.
You can access the protocol here. Come join us on this exciting journey!
• Congratulations to the team of Giorgio Gandaglia, Francesco Pellegrino, Bertrand De Meulder, Ayman Hijazy, Thomas Abbott, Asieh Golozar, Rossella Nicoletti, Juan Gomez-Rivas, Carl Steinbeisser, Susan Evans-Axelsson, Alberto Briganti, and James N’Dow on the recent publication of Research protocol for an observational health data analysis to assess the applicability of randomized controlled trials focusing on newly diagnosed metastatic prostate cancer using real-world data: PIONEER IMI’s “big data for better outcomes” program in the International Journal of Surgery Protocols.
• Congratulations to the team of Maria A Rujano, Jan-Willem Boiten, Christian Ohmann, Steve Canham, Sergio Contrino, Romain David, Jonathan Ewbank, Claudia Filippone, Claire Connellan, Ilse Custers, Rick van Nuland, Michaela Th Mayrhofer, Petr Holub, Eva García Álvarez, Emmanuel Bacry, Nigel Hughes, Mallory A Freeberg, Birgit Schaffhauser, Harald Wagener, Alex Sánchez-Pla, Guido Bertolini, Maria Panagiotopoulou on the recent publication of Sharing sensitive data in life sciences: an overview of centralized and federated approaches in Briefings in Bioinformatics.
• Congratulations to the team of Pedro Mateus, Justine Moonen, Magdalena Beran, Eva Jaarsma, Sophie M van der Landen, Joost Heuvelink, Mahlet Birhanu, Alexander Harms, Esther Bron, Frank J Wolters, Davy Cats, Hailiang Mei, Julie Oomens, Willemijn Jansen, Miranda T Schram, Andre Dekker, and Inigo Bermejo on the recent publication of Data harmonization and federated learning for multi-cohort dementia research using the OMOP common data model: A Netherlands consortium of dementia cohorts case study in the Journal of Biomedical Informatics.
• Congratulations to the team of Felix N Wirth, Hammam Abu Attieh, and Fabian Prasser on the recent publication of OHDSI-compliance: a set of document templates facilitating the implementation and operation of a software stack for real-world evidence generation in Frontiers in Medicine.
• Congratulations to the team of Shahim Essaid, Jeff Andre, Ian M Brooks, Katherine H Hohman, Madelyne Hull, Sandra L Jackson, Michael G Kahn, Emily M Kraus, Neha Mandadi, Amanda K Martinez, Joyce Y Mui, Bob Zambarano, and Andrey Soares on the publication of MENDS-on-FHIR: leveraging the OMOP common data model and FHIR standards for national chronic disease surveillance in the Journal of Biomedical Informatics.
• The next edition of the CBER BEST Seminar Series will be held Wednesday, June 26, at 11 am ET. Jenna Wong, Assistant Professor in the Department of Population Medicine at Harvard Medical School and Harvard Pilgrim Health Care Institute, will lead a session on Applying Machine Learning in Distributed Networks to Support Activities for Post-Market Surveillance of Medical Products: Opportunities, Challenges, and Considerations. The full schedule for the CBER BEST Seminar Series, including past presentations, is available here.
• If you are attending #DIA2024 in San Diego next week, there will be an OHDSI meetup on Tuesday, June 18, at 6 pm. Thanks to Davera Gabriel and Mui Van Zandt for leading this effort to connect, network and celebrate the many accomplishments of our community!
2024 OHDSI Global Symposium
• Registration is now open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
• Collaborator Showcase submissions are now being accepted, and all details about the event are available here. Submissions are due by Friday, June 21, at 8 pm ET. Notifications of acceptance will be sent out by Tuesday, Aug. 20.
Job Openings
• Aki Nishimura announced that Johns Hopkins University is seeking postdoctoral fellows. The fellows would work on methodological research in pharmaco-epidemiology to address medication and device utilization, effectiveness, and safety relevant to health, lung, and blood diseases. More information and application details are available here. If you are interested in having Aki Nishimura as a co-advisor, please include him on your application email at aki.nishimura@jhu.edu.
Slides
OHDSI Evidence Network Update | National Node Experiences: Belgium | Community Updates
Video Presentation
OHDSI Evidence Network Update (Network Study, Protocol, How To Join)
Europe Symposium Review
 The May 28 community call provided an opportunity for community members to collaborate on early research ideas for the OHDSI 2024 Global Symposium Collaborator Showcase.
The May 28 community call provided an opportunity for community members to collaborate on early research ideas for the OHDSI 2024 Global Symposium Collaborator Showcase.
The Collaborator Showcase has had a record number of submissions in each of the last two years, but many still wonder if their interests or current research fit into a showcase submission (hint: it probably does). Patrick Ryan shares thoughts on what type of research can benefit the community at the Global Symposium, which you can see at the bottom of this section.
Community Updates
• Congratulations to the team of Theresa Burkard, Kim López-Güell, Artem Gorbachev, Lucía Bellas, Annika Jödicke, Edward Burn, Maria de Ridder, Mees Mosseveld, Jasmine Gratton, Sarah Seager, Dina Vojinovic, Miguel Angel Mayer, Juan Manuel Ramírez-Anguita, Angela Leis Machín, Marek Oja, Raivo Kolde, Klaus Bonadt, Daniel Prieto-Alhambra, Chistian Reich, and Martí Català on the recent publication of Calculating daily dose in the Observational Medical Outcomes Partnership Common Data Model in Pharmacoepidemiology & Drug Safety.
• Congratulations to the team of Kayla Schiffer-Kane, Cong Liu, Tiffany J. Callahan, Casey Ta, Jordan G. Nestor, and Chunhua Weng on the publication of Converting OMOP CDM to phenopackets: A model alignment and patient data representation evaluation in the Journal of Biomedical Informatics.
• Congratulations to the team of Linying Zhang, Lauren Richter, Yixin Wang, Anna Ostropolets, Noémie Elhadad, David M. Blei, and George Hripcsak on the publication of Causal fairness assessment of treatment allocation with electronic health records in the Journal of Biomedical Informatics.
• Congratulations to the team of Minjung Han, Taehee Chang, Hae-Ryoung Chun, Suyoung Jo, Yeongchang Jo, Dong Han Yu, Sooyoung Yoo, and Sung-Il Cho on the publication of Symptoms and Conditions in Children and Adults up to 90 Days after SARS-CoV-2 Infection: A Retrospective Observational Study Utilizing the Common Data Model in the Journal of Clinical Medicine.
• The next edition of the CBER BEST Seminar Series will be held Wednesday, June 26, at 11 am ET. Jenna Wong, Assistant Professor in the Department of Population Medicine at Harvard Medical School and Harvard Pilgrim Health Care Institute, will lead a session on Applying Machine Learning in Distributed Networks to Support Activities for Post-Market Surveillance of Medical Products: Opportunities, Challenges, and Considerations. The full schedule for the CBER BEST Seminar Series, including past presentations, is available here.
• George Hripcsak hosted the previous edition of the CBER BEST Seminar Series last week and shared a presentation on Diagnosing Covariate Imbalance in Small-Cohort Studies. You can watch that video and check out his slides now.
• Applications are being accepted to join the 2024-25 Kheiron Cohort through June 1. This program is designed to onboard new contributors into OHDSI to empower them to become active contributors and maintainers. The goals are to provide career development, create a global perspective of the ecosystem, build cross-connections between projects and develop future leaders for OHDSI. You can apply for the upcoming cohort now!
• Linying Zhang is organizing a workshop on “AI for Reliable and Equitable Real-World Evidence Generation in Medicine” at the 22nd International Conference on Artificial Intelligence in Medicine (AIME), happening at Salt Lake City, Utah, on July 9-12, 2024. The workshop focuses on advancing the understanding and exploring the transformative role of artificial intelligence (AI) in analyzing real-world data (RWD) for real-world evidence (RWE) generation. George Hripcsak will be delivering the opening keynote speech. The workshop will also feature scientific presentations from authors with accepted papers or abstracts, and potentially a panel or debate. The intended length is 4 hours. We are calling for submissions of full papers, short papers, and poster abstracts! The submission deadline is May 31, 2024. More information is available on the workshop or AIME Conference homepages.
• The Center for Advanced Healthcare Research Informatics (CAHRI) at Tufts Medicine welcomes Peter Robinson, the Alexander von Humboldt Professor for AI at the Berlin Institute of Health at Charité, who will provide a talk on “The GA4GH Phenopacket Schema: A Standard for Computable Case Reports to Support Translational Genomic Research and Clinical Decision Support Software” on Thursday, May 30, at 11 am ET. Please contact Marty Alvarez at malvarez2@tuftsmedicalcenter.org for calendar invite or questions.
2024 OHDSI Global Symposium
• Registration is now open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
• Collaborator Showcase submissions are now being accepted, and all details about the event are available here. Submissions are due by Friday, June 21, at 8 pm ET. Notifications of acceptance will be sent out by Tuesday, Aug. 20.
Job Openings
• Aki Nishimura announced that Johns Hopkins University is seeking postdoctoral fellows. The fellows would work on methodological research in pharmaco-epidemiology to address medication and device utilization, effectiveness, and safety relevant to health, lung, and blood diseases. More information and application details are available here. If you are interested in having Aki Nishimura as a co-advisor, please include him on your application email at aki.nishimura@jhu.edu.
Slides
Video Presentation
Insights Into Submitting Research for the Collaborator Showcase
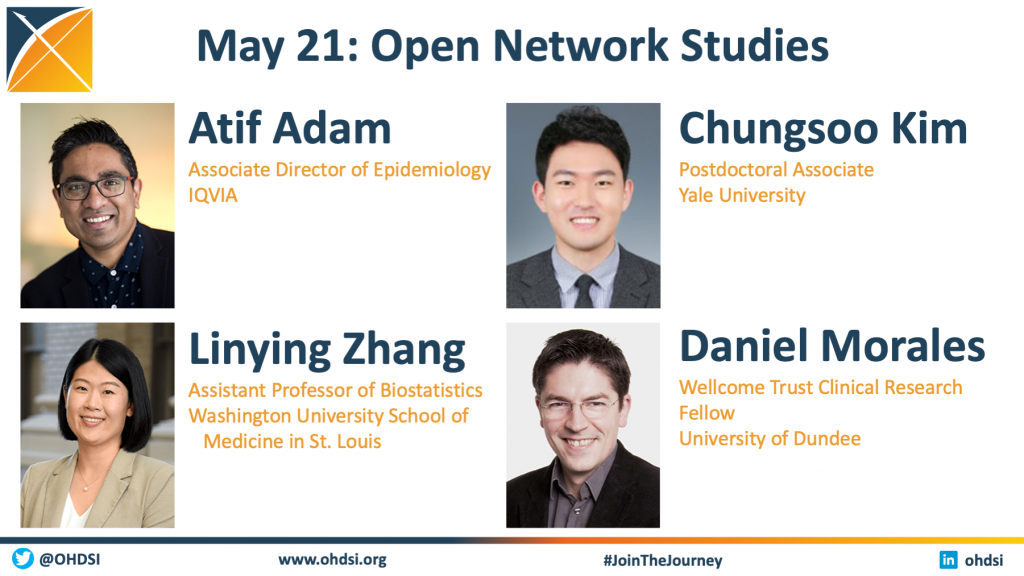 The May 21 community call featured four network studies that are taking place within the OHDSI community. We welcomed the following leads/representatives to discuss their studies:
The May 21 community call featured four network studies that are taking place within the OHDSI community. We welcomed the following leads/representatives to discuss their studies:
- Atif Adam, Associate Director of Epidemiology, IQVIA
- Linying Zhang, Assistant Professor of Biostatistics, Washington University School of Medicine in St. Louis
- Chungsoo Kim, Postdoctoral Associate, Yale University
- Daniel Morales, Wellcome Trust Clinical Research Fellow, University of Dundee
Community Updates
• Congratulations to the team of Phung-Anh Nguyen, Min-Huei Hsu, Tzu-Hao Chang, Hsuan-Chia Yang, Chih-Wei Huang, Chia-Te Liao, Christine Y. Lu, and Jason C. Hsu on the recent publication of Taipei Medical University Clinical Research Database: a collaborative hospital EHR database aligned with international common data standards in BMJ Health & Care Informatics.
• The next edition of the CBER BEST Initiative Seminar Series will be held Wednesday, May 22, at 11 am ET. We are excited to welcome George Hripcsak to share a presentation on Diagnosing Covariate Imbalance in Small-Cohort Studies. You can read about the talk, find the meeting link and watch past presentations on our CBER BEST Seminar Series homepage.
• Applications are being accepted to join the 2024-25 Kheiron Cohort. This program is designed to onboard new contributors into OHDSI to empower them to become active contributors and maintainers. The goals are to provide career development, create a global perspective of the ecosystem, build cross-connections between projects and develop future leaders for OHDSI. You can apply for the upcoming cohort now!
• Applications are now being accepted for the 2024 Maternal Health Data Science Fellowship, which is designed to empower clinical investigators to leverage emerging technologies for improved maternal and neonatal care while reducing morbidity and mortality. The program, which will include the components of career development, practice and networking, will train clinical investigators in observational research methods to enable them to conduct reproducible research and generate real-world evidence. More information, including application details, are now available, and the deadline to apply is May 22, 2024.
• Marissa Fiorella shared a forum post highlighting the upcoming Symposium on Risks and Opportunities of Artificial Intelligence in Pharmaceutical Medicine (AIPM), which will be held June 10-11 at Northeastern University, and will also have a Zoom option. Some OHDSI collaborators who are either speaking or helping organize the event are David Madigan, Asieh Golozar, David Sontag, Louisa Smith, and Marc Suchard. More details and registration information are available here.
• Linying Zhang is organizing a workshop on “AI for Reliable and Equitable Real-World Evidence Generation in Medicine” at the 22nd International Conference on Artificial Intelligence in Medicine (AIME), happening at Salt Lake City, Utah, on July 9-12, 2024. The workshop focuses on advancing the understanding and exploring the transformative role of artificial intelligence (AI) in analyzing real-world data (RWD) for real-world evidence (RWE) generation. George Hripcsakwill be delivering the opening keynote speech. The workshop will also feature scientific presentations from authors with accepted papers or abstracts, and potentially a panel or debate. The intended length is 4 hours. We are calling for submissions of full papers, short papers, and poster abstracts! The submission deadline is May 31, 2024. More information is available on the workshop or AIME Conference homepages.
• The Center for Advanced Healthcare Research Informatics (CAHRI) at Tufts Medicine welcomes Peter Robinson, the Alexander von Humboldt Professor for AI at the Berlin Institute of Health at Charité, who will provide a talk on “The GA4GH Phenopacket Schema: A Standard for Computable Case Reports to Support Translational Genomic Research and Clinical Decision Support Software” on Thursday, May 30, at 11 am ET. Please contact Marty Alvarez at malvarez2@tuftsmedicalcenter.org for calendar invite or questions.
• Registration is open for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
2024 OHDSI Global Symposium
• Registration is now open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
• Collaborator Showcase submissions are now being accepted, and all details about the event are available here. Submissions are due by Friday, June 21, at 8 pm ET. Notifications of acceptance will be sent out by Tuesday, Aug. 20.
OHDSI Social Showcase
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Sirius tool: Conversion of clinical study data into OMOP model and implementation of data quality monitoring of wearable sensor data (Vojtech Huser)
Tuesday — A Novel Approach to Matching Patients to Clinical Trials Using the OMOP Common Data Model (Jimmy John)
Wednesday — Improving the detection of behavioral health conditions through positive and unlabeled learning: opioid use disorder (Praveen Kumar)
Thursday — Quantifying Racial Disparities in Kidney Graft Failure Rates Using US Registry Data with Federated Learning Algorithms (Dazheng Zhang)
Job Openings
• Aki Nishimura announced that Johns Hopkins University is seeking postdoctoral fellows. The fellows would work on methodological research in pharmaco-epidemiology to address medication and device utilization, effectiveness, and safety relevant to health, lung, and blood diseases. More information and application details are available here. If you are interested in having Aki Nishimura as a co-advisor, please include him on your application email at aki.nishimura@jhu.edu.
Slides
ST-elevation myocardial infarction (STEMI) | Comparative Real-world Effectiveness of Medications for Opioid Use Disorder | Trials Replication through Observational study by Yonsei (TROY) | Community Updates
Video Presentations
ST-elevation myocardial infarction (STEMI)
Comparative Real-world Effectiveness of Medications for Opioid Use Disorder
Trials Replication through Observational study by Yonsei (TROY)
GLP1-RA and Thyroid Cancel (LEGEND-T2DM)
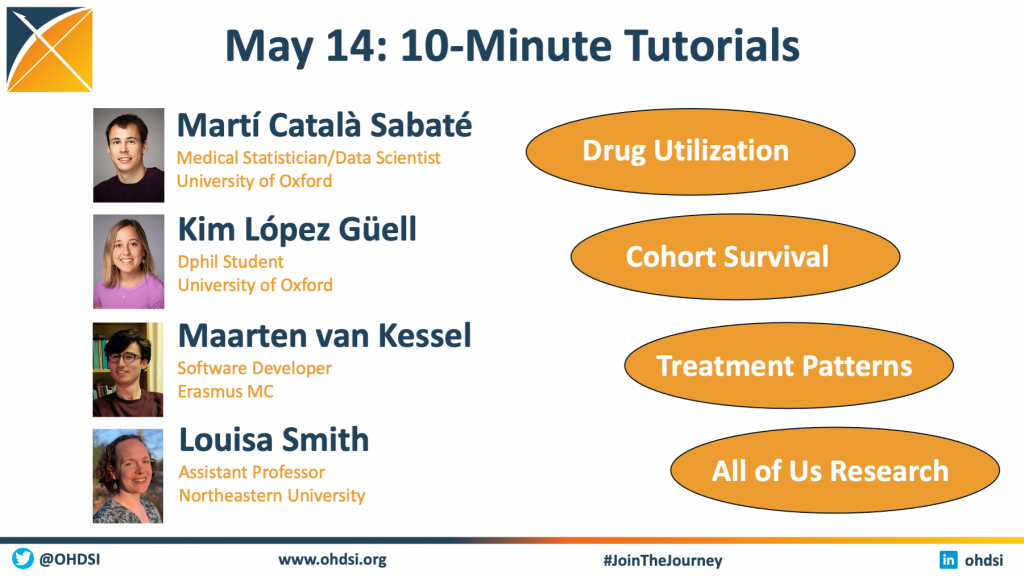 The May 14 community call featured a series of 10-minute tutorials, as four open-source development leaders provide introductions and brief live demonstrations on tools that can aid our community in observational healthcare research:
The May 14 community call featured a series of 10-minute tutorials, as four open-source development leaders provide introductions and brief live demonstrations on tools that can aid our community in observational healthcare research:
• Martí Català Sabaté, Medical Statistician/Data Scientist, University of Oxford (Drug Utilization)
• Kim López Güell, Dphil Student, University of Oxford (Cohort Survival)
• Maarten van Kessel, Software Developer, Erasmus MC (Treatment Pattens)
• Louisa Smith, Assistant Professor, Northeastern University (All of Us Research)
Community Updates
• The next edition of the CBER BEST Initiative Seminar Series will be held Wednesday, May 22, at 11 am ET. We are excited to welcome George Hripcsak to share a presentation on Diagnosing Covariate Imbalance in Small-Cohort Studies. You can read about the talk, find the meeting link and watch past presentations on our CBER BEST Seminar Series homepage.
• Applications are being accepted to join the 2024-25 Kheiron Cohort. This program is designed to onboard new contributors into OHDSI to empower them to become active contributors and maintainers. The goals are to provide career development, create a global perspective of the ecosystem, build cross-connections between projects and develop future leaders for OHDSI. You can apply for the upcoming cohort now!
• Applications are now being accepted for the 2024 Maternal Health Data Science Fellowship, which is designed to empower clinical investigators to leverage emerging technologies for improved maternal and neonatal care while reducing morbidity and mortality. The program, which will include the components of career development, practice and networking, will train clinical investigators in observational research methods to enable them to conduct reproducible research and generate real-world evidence. More information, including application details, are now available, and the deadline to apply is May 22, 2024.
• Marissa Fiorella shared a forum post highlighting the upcoming Symposium on Risks and Opportunities of Artificial Intelligence in Pharmaceutical Medicine (AIPM), which will be held June 10-11 at Northeastern University, and will also have a Zoom option. Some OHDSI collaborators who are either speaking or helping organize the event are David Madigan, Asieh Golozar, David Sontag, Louisa Smith, and Marc Suchard. More details and registration information are available here.
• Linying Zhang is organizing a workshop on “AI for Reliable and Equitable Real-World Evidence Generation in Medicine” at the 22nd International Conference on Artificial Intelligence in Medicine (AIME), happening at Salt Lake City, Utah, on July 9-12, 2024. The workshop focuses on advancing the understanding and exploring the transformative role of artificial intelligence (AI) in analyzing real-world data (RWD) for real-world evidence (RWE) generation. George Hripcsakwill be delivering the opening keynote speech. The workshop will also feature scientific presentations from authors with accepted papers or abstracts, and potentially a panel or debate. The intended length is 4 hours. We are calling for submissions of full papers, short papers, and poster abstracts! The submission deadline is May 31, 2024. More information is available on the workshop or AIME Conference homepages.
• Registration is open for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
2024 OHDSI Global Symposium
• Registration is now open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
• Collaborator Showcase submissions are now being accepted, and all details about the event are available here. Submissions are due by Friday, June 21, at 8 pm ET. Notifications of acceptance will be sent out by Tuesday, Aug. 20.
OHDSI Social Showcase
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Demonstrating Scalable Integration of Clinical, Translational, and Manufacturing Data to Explore Role of Manufacturing Approach in Driving Health Outcomes (Ben Smith)
Tuesday — Examining differential measurement error due to race, age, and sex in mental health disorders using PheValuator (Joel Swerdel)
Wednesday — Treatment pattern of osteoporosis in postmenopausal women using OMOP CDM (Dachung Boo)
Thursday — Unraveling the Mediating Role of Frailty: Understanding Health Care Utilization among Older Sexual and Gender Minority Adults in the All of Us Research Program (Chelsea Wong)
Friday — GUSTO Data Vault: Laying the foundations for an open science system with OMOP Data Catalogue (Cindy Ho)
Job Openings
• Aki Nishimura announced that Johns Hopkins University is seeking postdoctoral fellows. The fellows would work on methodological research in pharmaco-epidemiology to address medication and device utilization, effectiveness, and safety relevant to health, lung, and blood diseases. More information and application details are available here. If you are interested in having Aki Nishimura as a co-advisor, please include him on your application email at aki.nishimura@jhu.edu.
• Dani Prieto-Alhambra recently shared an opening for two Research Assistants in Health Data Sciences to join the Pharmaco- and Device epidemiology research group at the Botnar Research Centre, NDORMS, University of Oxford. In this role, you will contribute to the programming of analytical pipelines for the analysis of routinely collected data mapped to the OMOP Common Data Model. You will analyze real world data to address regulatory questions related to the prevalence/incidence of disease, use of medicines/vaccines, and the risks or benefits of medicines/vaccines or devices. You will prepare analytical packages to run a number of pre-specified analyses, contribute to wider project planning, including ideas for new research projects and gather, analyze, and present scientific data from a variety of sources. The application deadline is May 10. More information and an application link are available here.
Slides
Drug Utilization | All of Us Research | Community Updates
Video Presentations
Drug Utilization
Cohort Survival
Treatment Patterns
All of Us Research
 The May 7 community call featured a review of DevCon 2024, which served as an opportunity to connect our global open-source community and discuss ways we can collaborate and continue enhancing the future of OHDSI open-source software.
The May 7 community call featured a review of DevCon 2024, which served as an opportunity to connect our global open-source community and discuss ways we can collaborate and continue enhancing the future of OHDSI open-source software.
This session included several speakers and topics from the event, including:
• Paul Nagy, Johns Hopkins University (Open-Source Overview)
• Vishnu Chandrabalan, Lancaster University (OHDSI/OMOP – The hard way is the easy way)
• Roger Carlson, Spectrum Health (Moving OMOP to the Cloud with DBT and Snowflake)
• Lee Evans, LTS Computing LLC (Broadsea Update)
• Frank DeFalco, Janssen Research & Development (Technical Advisory Board (TAB) Update)
• Katy Sadowski, Boehringer Ingelheim (Kheiron Cohort Update)
This presentation is posted below. All recordings from the full DevCon 2024 session are available on the event homepage.
Community Updates
• Congratulations to the team of William G. Adams, Sarah Gasman, Ariel L. Beccia and Liza Fuentes on the publication of The Health Equity Explorer: An open-source resource for distributed health equity visualization and research across common data models in the Journal of Clinical and Translational Science.
• Congratulations to the team of Nhung TH Trinh, Annika M Jödicke, Martí Català, Núria Mercadé-Besora, Saeed Hayati, Angela Lupattelli, Daniel Prieto-Alhambra, and Hedvig ME Nordeng on the publication of Effectiveness of COVID-19 vaccines to prevent long COVID: data from Norway in The Lancet Respiratory Medicine.
• The latest edition of the OHDSI newsletter is now available, and it includes reflections on both the April Olympians and DevCon events, community updates, a collaborator showcase, publications and presentations from April, and plenty more. If you don’t receive the newsletter in your email, you can subscribe here.
• Montse Camprubi works at Synapse Research Management Partners. Currently the EHDEN community manager, she is leading the central coordination efforts between EHDEN Data Partners and certified SMES and EHDEN experts. In the latest edition of the collaborator spotlight, Montse discusses her background and career journey, recent highlights and future plans in EHDEN, the upcoming OHDSI Europe Symposium, and plenty more.
• Applications are being accepted to join the 2024-25 Kheiron Cohort. This program is designed to onboard new contributors into OHDSI to empower them to become active contributors and maintainers. The goals are to provide career development, create a global perspective of the ecosystem, build cross-connections between projects and develop future leaders for OHDSI. You can apply for the upcoming cohort now!
• Applications are now being accepted for the 2024 Maternal Health Data Science Fellowship, which is designed to empower clinical investigators to leverage emerging technologies for improved maternal and neonatal care while reducing morbidity and mortality. The program, which will include the components of career development, practice and networking, will train clinical investigators in observational research methods to enable them to conduct reproducible research and generate real-world evidence. More information, including application details, are now available, and the deadline to apply is May 22, 2024.
• Marissa Fiorella shared a forum post highlighting the upcoming Symposium on Risks and Opportunities of Artificial Intelligence in Pharmaceutical Medicine (AIPM), which will be held June 10-11 at Northeastern University, and will also have a Zoom option. Some OHDSI collaborators who are either speaking or helping organize the event are David Madigan, Asieh Golozar, David Sontag, Louisa Smith, and Marc Suchard. More details and registration information are available here.
• Linying Zhang is organizing a workshop on “AI for Reliable and Equitable Real-World Evidence Generation in Medicine” at the 22nd International Conference on Artificial Intelligence in Medicine (AIME), happening at Salt Lake City, Utah, on July 9-12, 2024. The workshop focuses on advancing the understanding and exploring the transformative role of artificial intelligence (AI) in analyzing real-world data (RWD) for real-world evidence (RWE) generation. George Hripcsakwill be delivering the opening keynote speech. The workshop will also feature scientific presentations from authors with accepted papers or abstracts, and potentially a panel or debate. The intended length is 4 hours. We are calling for submissions of full papers, short papers, and poster abstracts! The submission deadline is May 31, 2024. More information is available on the workshop or AIME Conference homepages.
• Registration is open for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
2024 OHDSI Global Symposium
• Registration is now open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
• Collaborator Showcase submissions are now being accepted, and all details about the event are available here. Submissions are due by Friday, June 21, at 8 pm ET. Notifications of acceptance will be sent out by Tuesday, Aug. 20.
OHDSI Social Showcase
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Jackalope Plus: AI-Enhanced Solution for Mapping Unmappable Concepts (Denys Kaduk)
Tuesday — Framework and Implementation of an OMOP-Oriented Clinical Data Warehouse Using Databricks (Jared Houghtaling)
Wednesday — Mother-Infant Linked Data: Methodology, Case Studies, and Cohort Development for Investigating Prenatal Exposure and Neonatal Outcomes (Jill Hardin)
Thursday — Integrating ATLAS Cohorts with DICOM Images and ECG Waveforms to Enrich Real-World Evidence Research (Boudewijn Aasman)
Friday — Real-world Effectiveness of BNT162b2 in Children and Adolescents in Preventing Infection and Severe Diseases with SARS-CoV-2 During the Delta and Omicron Periods (Qiong Wu)
Job Openings
• Ross Williams shared that the Erasmus MC Department of Medical Informatics is looking for a PhD student to work on cutting-edge health AI and data science topics. This person will be responsible for the research on using federated data networks to improve best practices around the development and validation of prediction models. You will lead and contribute to projects conducting methodological research within the field of machine learning in healthcare. More information and the application link are available here. The application deadline is May 13.
• Aki Nishimura announced that Johns Hopkins University is seeking postdoctoral fellows. The fellows would work on methodological research in pharmaco-epidemiology to address medication and device utilization, effectiveness, and safety relevant to health, lung, and blood diseases. More information and application details are available here. If you are interested in having Aki Nishimura as a co-advisor, please include him on your application email at aki.nishimura@jhu.edu.
• Dani Prieto-Alhambra recently shared an opening for two Research Assistants in Health Data Sciences to join the Pharmaco- and Device epidemiology research group at the Botnar Research Centre, NDORMS, University of Oxford. In this role, you will contribute to the programming of analytical pipelines for the analysis of routinely collected data mapped to the OMOP Common Data Model. You will analyze real world data to address regulatory questions related to the prevalence/incidence of disease, use of medicines/vaccines, and the risks or benefits of medicines/vaccines or devices. You will prepare analytical packages to run a number of pre-specified analyses, contribute to wider project planning, including ideas for new research projects and gather, analyze, and present scientific data from a variety of sources. The application deadline is May 10. More information and an application link are available here.
Slides
DevCon Overview | Presentations: Chandrabalan / Carlson | Community Updates
Video Presentation
The conclusion of the April Olympians collaborative activity includes thanks to the many community members who took part, and an interactive game that highlights how to use the newly developed Themis conventions homepage. This session is led by Clair Blacketer, Director, Observational Health Data Analytics, Janssen Research & Development.
Community Updates
• Congratulations to the team of Martin Baumgartner, Karl Kreiner, Aaron Lauschensky, Bernhard Jammerbund, Klaus Donsa, Dieter Hayn, Fabian Wiesmüller, Lea Demelius, Robert Modre-Osprian, Sabrina Neururer, Gerald Slamanig, Sarah Prantl, Luca Brunelli, Bernhard Pfeifer, Gerhard Pölzl, and Günter Schreier on the publication of Health data space nodes for privacy-preserving linkage of medical data to support collaborative secondary analyses in Frontiers in Medicine.
• Congratulations to the team of Chungsoo Kim, Dong Han Yu, Hyeran Baek, Jaehyeong Cho, Seng Chan You, and Rae Woong Park on the publication of Data Resource Profile: Health Insurance Review and Assessment Service Covid-19 Observational Medical Outcomes Partnership (HIRA Covid-19 OMOP) database in South Korea in the International Journal of Epidemiology.
• Congratulations to the team of Markus Falgenhauer, Aaron Lauschensky, Karl Kreiner, Stefan Beyer, Kristina Reiter, Andreas Ziegl, Robert Modre-Osprian, Bernhard Pfeifer, Sabrina Neururer, Susanne Krestan, Hanna Wagner, Andreas Huber, Sandra Plaikner, Sarah Kuppelwieser, Martin Widschwendter, and Günter Schreier on the publication of Towards an Electronic Health Prevention Record Based on HL7 FHIR and the OMOP Common Data Model in Volume 313 of Studies in Health Technology and Informatics.
• Congratulations to the team of Evgeniy Krastev, Emanuil Markov, Simeon Abanos, Ralitsa Krasteva, and Dimitar Tcharaktchiev on the publication of Mapping the Bulgarian Diabetes Register to OMOP CDM: Application Results in Volume 313 of Studies in Health Technology and Informatics.
• DevCon 2024 served as an opportunity to connect our global open-source community and discuss ways we can collaborate and continue enhancing the future of OHDSI open-source software. The full agenda from the event is posted below, and recordings are available on the event homepage.
• Applications are being accepted to join the 2024-25 Kheiron Cohort. This program is designed to onboard new contributors into OHDSI to empower them to become active contributors and maintainers. The goals are to provide career development, create a global perspective of the ecosystem, build cross-connections between projects and develop future leaders for OHDSI. You can apply for the upcoming cohort now!
• Applications are now being accepted for the 2024 Maternal Health Data Science Fellowship, which is designed to empower clinical investigators to leverage emerging technologies for improved maternal and neonatal care while reducing morbidity and mortality. The program, which will include the components of career development, practice and networking, will train clinical investigators in observational research methods to enable them to conduct reproducible research and generate real-world evidence. More information, including application details, are now available, and the deadline to apply is May 15, 2024.
• Linying Zhang is organizing a workshop on “AI for Reliable and Equitable Real-World Evidence Generation in Medicine” at the 22nd International Conference on Artificial Intelligence in Medicine (AIME), happening at Salt Lake City, Utah, on July 9-12, 2024. The workshop focuses on advancing the understanding and exploring the transformative role of artificial intelligence (AI) in analyzing real-world data (RWD) for real-world evidence (RWE) generation. George Hripcsakwill be delivering the opening keynote speech. The workshop will also feature scientific presentations from authors with accepted papers or abstracts, and potentially a panel or debate. The intended length is 4 hours. We are calling for submissions of full papers, short papers, and poster abstracts! The submission deadline is May 31, 2024. More information is available on the workshop or AIME Conference homepages.
• Registration is open for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
2024 OHDSI Global Symposium
• Registration is now open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
• Collaborator Showcase submissions are now being accepted, and all details about the event are available here. Submissions are due by Friday, June 21, at 8 pm ET. Notifications of acceptance will be sent out by Tuesday, Aug. 20.
OHDSI Social Showcase
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Opportunity and Challenge of Implementing the OHDSI System in Indonesia (Dian Tri Wiyanti)
Tuesday — HowOften: Large Scale Incidence Rate Calculation of Every Side Effect for Every Drug (Elise Ruan)
Wednesday — Agreement between measurement and diagnosis-based phenotype algorithms (Azza Shoaibi)
Thursday — Comparing Patient Self-Reported Symptoms with SNOMED/ICD-10-CM Codes at Primary Care Visits (Victor Castro)
Friday — Using the Informatics for Integrating Biology and the Bedside Platform to Query OMOP Data in the OHDSI Ecosystem (Jeffrey Klann)
Job Openings
• Dani Prieto-Alhambra recently shared an opening for two Research Assistants in Health Data Sciences to join the Pharmaco- and Device epidemiology research group at the Botnar Research Centre, NDORMS, University of Oxford. In this role, you will contribute to the programming of analytical pipelines for the analysis of routinely collected data mapped to the OMOP Common Data Model. You will analyze real world data to address regulatory questions related to the prevalence/incidence of disease, use of medicines/vaccines, and the risks or benefits of medicines/vaccines or devices. You will prepare analytical packages to run a number of pre-specified analyses, contribute to wider project planning, including ideas for new research projects and gather, analyze, and present scientific data from a variety of sources. The application deadline is May 10. More information and an application link are available here.
• Alex Asiimwe shared an opening for a Director, RWE – Data Science at Gilead. As a RWE Data Scientist (OMOP/OHDSI), you will play a crucial role in designing, implementing, and maintaining healthcare data solutions using the OHDSI framework. This position offers an exciting opportunity to contribute to advancements in health informatics and research.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
Slides
April Olympians Conclusion | Community Updates
Video Presentation
 The April 23 community call focused on the CDM and Themis Process. This call provided historical and current information about both the CDM and Themis workgroups, including their mission and processes. Presentations were made by our April Olympians co-leads:
The April 23 community call focused on the CDM and Themis Process. This call provided historical and current information about both the CDM and Themis workgroups, including their mission and processes. Presentations were made by our April Olympians co-leads:
– Clair Blacketer, Director, Observational Health Data Analytics, Janssen Research & Development
– Melanie Philofsky, Senior Business Analyst and Project Manager, Odysseus Data Services, Inc.
This session also included our fourth update on the April Olympians community activity. You can watch the full presentation below.
Community Updates
• Congratulations to the team of Roger Ward, Christine Mary Hallinan, David Ormiston-Smith, Christine Chidgey, and Dougie Boyle on the publication of The OMOP common data model in Australian primary care data: Building a quality research ready harmonised dataset in PLOS One.
• Congratulations to the team of Christian Gulden, Philipp Macho, Ines Reinecke, Cosima Strantz, Hans-Ulrich Prokosch, and Romina Blasini on the publication of recruIT: A cloud-native clinical trial recruitment support system based on Health Level 7 Fast Healthcare Interoperability Resources (HL7 FHIR) and the Observational Medical Outcomes Partnership Common Data Model (OMOP CDM) in Computers in Biology and Medicine.
• Congratulations to the team of Giorgio Gandaglia, Francesco Pellegrino, Asieh Golozar, Bertrand De Meulder, Thomas Abbott, Ariel Achtman, Muhammad Imran Omar, Thamir Alshammari, Carlos Areia, Alex Asiimwe, Katharina Beyer, Anders Bjartell, Riccardo Campi, Philip Cornford, Thomas Falconer, Qi Feng, Mengchun Gong, Ronald Herrera, Nigel Hughes, Tim Hulsen, Adam Kinnaird, Lana Y.H. Lai, Gianluca Maresca, Nicolas Mottet, Marek Oja, Peter Prinsen, Christian Reich, Sebastiaan Remmers, Monique J. Roobol, Vasileios Sakalis, Sarah Seager, Emma J. Smith, Robert Snijder, Carl Steinbeisser, Nicolas H. Thurin, Ayman Hijazy, Kees van Bochove, Roderick C.N. Van den Bergh, Mieke Van Hemelrijck, Peter-Paul Willemse, Andrew E. Williams, Nazanin Zounemat Kermani, Susan Evans-Axelsson, Alberto Briganti, James N’Dow, on behalf of the PIONEER Consortium on the publication of Clinical Characterization of Patients Diagnosed with Prostate Cancer and Undergoing Conservative Management: A PIONEER Analysis Based on Big Data in European Urology.
• Applications are now being accepted for the 2024 Maternal Health Data Science Fellowship, which is designed to empower clinical investigators to leverage emerging technologies for improved maternal and neonatal care while reducing morbidity and mortality. The program, which will include the components of career development, practice and networking, will train clinical investigators in observational research methods to enable them to conduct reproducible research and generate real-world evidence. More information, including application details, are now available, and the deadline to apply is May 15, 2024.
 • The third annual OHDSI DevCon will be held virtually on Friday, April 26, from 9 am-3 pm ET. Join leaders from our Open-Source Community for a day to both welcome and inform both new and veteran developers within the OHDSI Community. More details on the agenda will be posted when available.
• The third annual OHDSI DevCon will be held virtually on Friday, April 26, from 9 am-3 pm ET. Join leaders from our Open-Source Community for a day to both welcome and inform both new and veteran developers within the OHDSI Community. More details on the agenda will be posted when available.
• Registration is now OPEN for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
2024 OHDSI Global Symposium
• Registration is now open for the 2024 Global Symposium, which will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, N.J., USA. The three-day event will feature tutorials on Day 1, plenaries and the collaborator showcase on Day 2, and workgroup activities on Day 3.
• Day 1 will open with a single tutorial in the morning: An Introduction to the Journey from Data to Evidence using OHDSI. There will be four advanced tutorials during the afternoon: An Introduction to the Journey from Data to Evidence using OHDSI; Developing and Evaluating Your Extract, Transform, Load (ETL) Process to the OMOP Common Data Model; So, You Think You Want To Run an OHDSI Network Study?; and Using the OHDSI Standardized Vocabularies for Research. You can select your tutorials during the registration process.
• Collaborator Showcase submissions are now being accepted, and all details about the event are available here. Submissions are due by Friday, June 21, at 8 pm ET. Notifications of acceptance will be sent out by Tuesday, Aug. 20.
OHDSI Social Showcase
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Enhancing Data Quality Management: Introducing Capture and Cleanse Modes to the Data Quality Dashboard (Frank DeFalco)
Tuesday — Making OMOP Happen: An Implementation Science Approach (Maya Younoszai)
Wednesday — Evaluation of Study Execution using Large-Scale Analytics: A Machine Learning Approach to Assess Pre-Exposure Prophylaxis (PrEP) Utilization in the Real-World (Nag Mani)
Thursday — Validation and Comparison of Frailty Indexes: An OHDSI Network Study (Chen Yanover)
Friday — Broadsea 3.0: “BROADening the ohdSEA” (Ajit Londhe)
Job Openings
• Dani Prieto-Alhambra recently shared an opening for two Research Assistants in Health Data Sciences to join the Pharmaco- and Device epidemiology research group at the Botnar Research Centre, NDORMS, University of Oxford. In this role, you will contribute to the programming of analytical pipelines for the analysis of routinely collected data mapped to the OMOP Common Data Model. You will analyze real world data to address regulatory questions related to the prevalence/incidence of disease, use of medicines/vaccines, and the risks or benefits of medicines/vaccines or devices. You will prepare analytical packages to run a number of pre-specified analyses, contribute to wider project planning, including ideas for new research projects and gather, analyze, and present scientific data from a variety of sources. The application deadline is May 10. More information and an application link are available here.
• Alex Asiimwe shared an opening for a Director, RWE – Data Science at Gilead. As a RWE Data Scientist (OMOP/OHDSI), you will play a crucial role in designing, implementing, and maintaining healthcare data solutions using the OHDSI framework. This position offers an exciting opportunity to contribute to advancements in health informatics and research.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
Slides
CDM Process | Themis Process | Community Updates
Video Presentation
 The April 16 community call focused on Tools to Evaluate ETL. We welcomed two community leaders to lead our main session:
The April 16 community call focused on Tools to Evaluate ETL. We welcomed two community leaders to lead our main session:
– Frank DeFalco, Senior Director, Observational Health Data Analytics at Janssen Research & Development
– Katy Sadowski, Senior Associate Director, Boehringer Ingelheim
This session also included our third update on the April Olympians community activity. Both presentations are available below.
Community Updates
• Congratulations to the team of Nhung TH Trinh, Annika M Jödicke, Martí Català, Núria Mercadé-Besora, Saeed Hayati, Angela Lupattelli, Daniel Prieto-Alhambra, and Hedvig ME Nordeng on the publication of Effectiveness of COVID-19 vaccines to prevent long COVID: data from Norway in The Lancet Respiratory Medicine.
• Congratulations to the team of Guy Tsafnat, Rachel Dunscombe, Davera Gabriel, Grahame Grieve, and Christian Reich on the publication of Converge or Collide? Making Sense of a Plethora of Open Data Standards in Health Care in the Journal of Medical Internet Research.
• Congratulations to the team of Ailbhe Lawlor, Carol Lin, Juan Gómez Rivas, Laura Ibáñez, Pablo Abad López, Peter-Paul Willemse, Muhammad Imran Omar, Sebastiaan Remmers, Philip Cornford, Pawel Rajwa, Rossella Nicoletti, Giorgio Gandaglia, Jeremy Yuen-Chun Teoh, Jesús Moreno Sierra, Asieh Golozar, Anders Bjartell, Susan Evans-Axelsson, James N’Dow, Jihong Zong, Maria J. Ribal, Monique J. Roobol, Mieke Van Hemelrijck, Katharina Beyer, on behalf of the PIONEER Consortium on the publication of Predictive Models for Assessing Patients’ Response to Treatment in Metastatic Prostate Cancer: A Systematic Review in European Urology Open Science.
• Applications are now being accepted for the 2024 Maternal Health Data Science Fellowship, which is designed to empower clinical investigators to leverage emerging technologies for improved maternal and neonatal care while reducing morbidity and mortality. The program, which will include the components of career development, practice and networking, will train clinical investigators in observational research methods to enable them to conduct reproducible research and generate real-world evidence. More information, including application details, are now available, and the deadline to apply is May 15, 2024.
• The CBER BEST Initiative Seminar Series returns Wednesday, April 17 (11 am – 12 pm ET) as 2021 Titan Award honoree Yong Chen presents his research on“Real-World Effectiveness of BNT162b2 Against Infection and Severe Diseases in Children and Adolescents: causal inference under misclassification in treatment status.” This series is open to anybody: Calendar invite to CBER BEST Seminar.
 • The third annual OHDSI DevCon will be held virtually on Friday, April 26, from 9 am-3 pm ET. Join leaders from our Open-Source Community for a day to both welcome and inform both new and veteran developers within the OHDSI Community. More details on the agenda will be posted when available.
• The third annual OHDSI DevCon will be held virtually on Friday, April 26, from 9 am-3 pm ET. Join leaders from our Open-Source Community for a day to both welcome and inform both new and veteran developers within the OHDSI Community. More details on the agenda will be posted when available.
• The 2024 OHDSI Global Symposium will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, NJ. The tentative symposium format will feature tutorials on Oct. 22, plenaries and the collaborator showcase on Oct. 23, and workgroup activities on Oct. 24. Registration has not opened yet.
• Registration is now OPEN for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
• Atif Adam announced an opportunity for collaboration around a new network study focusing on Acute ST-Elevation Myocardial Infarction (STEMI). The study intends to deepen the understanding of STEMI patients’ characteristics and identify incidence rates across multiple real-world data datasets. More details and information on how to get involved are available within the forum post.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Augmenting the National COVID Cohort Collaborative (N3C) Dataset with Medicare and Medicaid (CMS) Data, Secure and Deidentified Clinical Dataset (Stephanie Hong)
Tuesday — The Feasibility of Clinical Quality Language (CQL) Based Digital Quality Measures (dQMs) Implementation to OMOP CDM (Emir Amaro Syailendra)
Wednesday — Using Cohort Diagnostics to Assess the Phenotypic Data Quality in All of Us Research Program (Lina Sulieman)
Thursday — Demonstration of the OHDSI phenotype library (Gowtham Rao)
Friday — Large variety Country size RWD data-lake (Guy Livne)
Job Openings
• Alex Asiimwe shared an opening for a Director, RWE – Data Science at Gilead. As a RWE Data Scientist (OMOP/OHDSI), you will play a crucial role in designing, implementing, and maintaining healthcare data solutions using the OHDSI framework. This position offers an exciting opportunity to contribute to advancements in health informatics and research.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
Slides
ACHILLES/ARES | DataQualityDashboard | Community Updates
Video Presentation
Tools to Evaluate ETL
April Olympians Update
 The April 9 community call focused on Vocabulary Techniques for ETL. This call also included our second April Olympians update (Clair Blacketer and Melanie Philofsky), as well as the first OKR presentation from the newly formed Rehabilitation Workgroup (Ruud Selles and Esther Janssen).
The April 9 community call focused on Vocabulary Techniques for ETL. This call also included our second April Olympians update (Clair Blacketer and Melanie Philofsky), as well as the first OKR presentation from the newly formed Rehabilitation Workgroup (Ruud Selles and Esther Janssen).
We welcomed four community leaders to present on Vocabulary Techiques for ETL:
– Dmitry Dymshyts, Associate Director, Janssen Research & Development
– Tanya Skugarevskaya, Vocabulary Team, Odysseus Data Services, Inc.
– Anna Ostropolets, Associate Director, Janssen Research & Development
– Alexander Davydov, Director, Lead of Medical Ontologies, Odysseus Data Services, Inc.
Community Updates
• Congratulations to the team of Pawel Rajwa, Angelika Borkowetz, Thomas Abbott, Andrea Alberti, Anders Bjartell, James T. Brash, Riccardo Campi, Andrew Chilelli, Mitchell Conover, Niculae Constantinovici, Eleanor Davies, Bertrand De Meulder, Sherrine Eid, Mauro Gacci, Asieh Golozar, Haroon Hafeez, Samiul Haque, Ayman Hijazy, Tim Hulsen, Andreas Josefsson, Sara Khalid, Raivo Kolde, Daniel Kotik, Samu Kurki, Mark Lambrecht, Chi-Ho Leung, Julia Moreno, Rossella Nicoletti, Daan Nieboer, Marek Oja, Soundarya Palanisamy, Peter Prinsen, Christian Reich, Giulio Raffaele Resta, Maria J Ribal, Juan Gómez Rivas, Emma Smith, Robert Snijder, Carl Steinbeisser, Frederik Vandenberghe, Philip Cornford, Susan Evans-Axelsson, James N’Dow, and Peter-Paul M Willemse on the publication of Research Protocol for an Observational Health Data Analysis on the Adverse Events of Systemic Treatment in Patients with Metastatic Hormone-sensitive Prostate Cancer: Big Data Analytics Using the PIONEER Platform in European Urology Open Science.
• The CBER BEST Initiative Seminar Series returns Wednesday, April 17 (11 am – 12 pm ET) as 2021 Titan Award honoree Yong Chen presents his research on“Real-World Effectiveness of BNT162b2 Against Infection and Severe Diseases in Children and Adolescents: causal inference under misclassification in treatment status.” This series is open to anybody: Calendar invite to CBER BEST Seminar.
• The latest OHDSI newsletter is now available. This newsletter includes information on the recent standardized vocabularies release, a preview of the April Olympians collaborative activity, the latest collaborator spotlight on Melanie Philofsky, the monthly videocast, links to the nine published studies from the OHDSI community in March, and plenty more. If you don’t receive the newsletter in your inbox, you can subscribe here.
• Melanie Philofsky is a Senior Business & Data Analyst with Odysseus Data Services, Inc. She is responsible for the harmonization of various healthcare data sources into the OMOP Common Data Model to support research endeavors. Her areas of expertise include clinical informatics, data analysis, data quality, ETL conversions, EHR data, the OMOP CDM and data modeling of new domains. Melanie was the 2022 Titan Award honoree for Contributions in Data Standards. In the latest edition of the Collaborator Spotlight, she discusses her career journey, her work with the Healthcare Systems and Themis workgroups, plans for the April Olympians Collab-a-thon, and more!
• The HADES-wide release 2024Q1 has been released. This is a snapshot of the HADES packages and their dependencies, thoroughly tested and confirmed to be mutually compatible. It is intended to be a stable environment for studies and execution engines. Currently, the release is only available as an renv lock file 3, but folks are working on containers as well.
• The third annual OHDSI DevCon will be held virtually on Friday, April 26, from 9 am-3 pm ET. Join leaders from our Open-Source Community for a day to both welcome and inform both new and veteran developers within the OHDSI Community. More details on the agenda will be posted when available.
• The 2024 OHDSI Global Symposium will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, NJ. The tentative symposium format will feature tutorials on Oct. 22, plenaries and the collaborator showcase on Oct. 23, and workgroup activities on Oct. 24. Registration has not opened yet.
• Registration is now OPEN for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
• Atif Adam announced an opportunity for collaboration around a new network study focusing on Acute ST-Elevation Myocardial Infarction (STEMI). The study intends to deepen the understanding of STEMI patients’ characteristics and identify incidence rates across multiple real-world data datasets. More details and information on how to get involved are available within the forum post.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Implementing the OMOP common data model in an NHS Trust using DBT (Quinta Ashcroft)
Tuesday — Mining Data Outside the Box: Internet as a New Source for Common Data Model (Min-Gyu Kim)
Wednesday — Forecasting Daily Incidence of Respiratory Symptoms: A Comparative Study on Time Series Models using OMOP-CDM in South Korea (Min Ho An)
Thursday — Observational Research in Dentistry: A Scoping Review (Robert Koski)
Friday — Integration of Scalable Natural Language Processing to the Atlas Cohort Building Workflow (Pavan Parimi)
Job Openings
• Alex Asiimwe shared an opening for a Director, RWE – Data Science at Gilead. As a RWE Data Scientist (OMOP/OHDSI), you will play a crucial role in designing, implementing, and maintaining healthcare data solutions using the OHDSI framework. This position offers an exciting opportunity to contribute to advancements in health informatics and research.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
Slides
Vocabulary Techniques for ETL | Future Directions in Vocabularies | Community Updates
Video Presentation
The April 2 (11 am ET) community call featured our first “April Olympians” session. Event co-leads Clair Blacketer and Melanie Philofsky provided the first update, including examples of what the ‘hunters’ and ‘writers’ will do during the month, and details on how to get involved throughout the month. Anton Ivanov also joined to discuss the Perseus ETL tool.
Community Updates
• Congratulations to the team of Kipyo Kim, Ji-Eun Kim, Jae Ho Kim, Seong Hee Ahn, Chai Young Jung, Seun Deuk Hwang, Seoung Woo Lee and Joon Ho Song on the publication of Real-world evidence of constipation and laxative use in the Korean population with chronic kidney disease from a common data model in Scientific Reports.
• Congratulations to the team of Cindy Cai, Akihiko Nishimura, Mary Bowring, Erik Westlund, Diep Tran, Jia Ng, Paul Nagy, Michael Cook, Jody-Ann McLeggon, Scott DuVall, Michael Matheny, Asieh Golozar, Anna Ostropolets, Evan Minty, Priya Desai, Fan Bu, Brian Toy, Michelle Hribar, Thomas Falconer, Linying Zhang, Laurence Lawrence-Archer, Michael Boland, Kerry Goetz, Nathan Hall, Azza Shoaibi, Jenna Reps, Anthony Sena, Clair Blacketer, Joel Swerdel, Kenar Jhaveri, Edward Lee, Zachary Gilbert, Scott Zeger, Deidra Crews, Marc Suchard, George Hripcsak, and Patrick Ryan on the publication of Similar risk of kidney failure among patients with blinding diseases who receive ranibizumab, aflibercept, and bevacizumab: an OHDSI Network Study in Ophthalmology Retina.
• Congratulations to the team of Joshua Ide, Azza Shoaibi, Kerstin Wagner, Rachel Weinstein, Kathleen E. Boyle and Andrew Myers on the publication of Patterns of Comorbidities and Prescribing and Dispensing of Non-steroidal Anti-inflammatory Drugs (NSAIDs) Among Patients with Osteoarthritis in the USA: Real-World Study in Drugs & Aging.
• Congratulations to the team of Jens Weidner, Ingmar Glauche, Ulf Manuwald, Ivana Kern, Ines Reinecke, Franziska Bathelt, Makan Amin, Fan Dong, Ulrike Rothe, and Joachim Kugler on the publication of Correlation of Socioeconomic and Environmental Factors With Incidence of Crohn Disease in Children and Adolescents: Systematic Review and Meta-Regression in JMR Public Health and Surveillance.
• Congratulations to the team of Valerie van Baalen, Eva-Maria Didden, Daniel Rosenberg, Kristina Bardenheuer, Michel van Speybroeck, and Monika Brand on the publication of Increase transparency and reproducibility of real-world evidence in rare diseases through disease-specific Federated Data Networks in Pharmacoepidemiology & Drug Safety.
• The CBER BEST Initiative Seminar Series returns Wednesday, April 17 (11 am – 12 pm ET) as 2021 Titan Award honoree Yong Chen presents his research on“Real-World Effectiveness of BNT162b2 Against Infection and Severe Diseases in Children and Adolescents: causal inference under misclassification in treatment status.” This series is open to anybody: Calendar invite to CBER BEST Seminar.
• The latest OHDSI newsletter is now available. This newsletter includes information on the recent standardized vocabularies release, a preview of the April Olympians collaborative activity, the latest collaborator spotlight on Melanie Philofsky, the monthly videocast, links to the nine published studies from the OHDSI community in March, and plenty more. If you don’t receive the newsletter in your inbox, you can subscribe here.
• Melanie Philofsky is a Senior Business & Data Analyst with Odysseus Data Services, Inc. She is responsible for the harmonization of various healthcare data sources into the OMOP Common Data Model to support research endeavors. Her areas of expertise include clinical informatics, data analysis, data quality, ETL conversions, EHR data, the OMOP CDM and data modeling of new domains. Melanie was the 2022 Titan Award honoree for Contributions in Data Standards. In the latest edition of the Collaborator Spotlight, she discusses her career journey, her work with the Healthcare Systems and Themis workgroups, plans for the April Olympians Collab-a-thon, and more!
• The third annual OHDSI DevCon will be held virtually on Friday, April 26, from 9 am-3 pm ET. Join leaders from our Open-Source Community for a day to both welcome and inform both new and veteran developers within the OHDSI Community. More details on the agenda will be posted when available.
• The 2024 OHDSI Global Symposium will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, NJ. The tentative symposium format will feature tutorials on Oct. 22, plenaries and the collaborator showcase on Oct. 23, and workgroup activities on Oct. 24. Registration has not opened yet.
• Registration is now OPEN for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
• Atif Adam announced an opportunity for collaboration around a new network study focusing on Acute ST-Elevation Myocardial Infarction (STEMI). The study intends to deepen the understanding of STEMI patients’ characteristics and identify incidence rates across multiple real-world data datasets. More details and information on how to get involved are available within the forum post.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Integrating clinical and laboratory research data using the OMOP CDM (Edward Frankenberger)
Tuesday — A new route of administration hierarchy derived from dose forms supporting standardised drug dose calculations (Theresa Burkard)
Wednesday — Bayesian Evidence Synthesis with Bias Correction (Louisa Smith)
Thursday — Save Our Sisyphus Challenge: Lessons learned from Strategus execution on the OHDSI Network (Anthony Sena)
Friday — Incorporating Real-World Data Research in Training First-Year Medical Students Using OHDSI OMOP and Atlas tools (Pavel Goriacko)
Job Openings
• Alex Asiimwe shared an opening for a Director, RWE – Data Science at Gilead. As a RWE Data Scientist (OMOP/OHDSI), you will play a crucial role in designing, implementing, and maintaining healthcare data solutions using the OHDSI framework. This position offers an exciting opportunity to contribute to advancements in health informatics and research.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
• Nathan Hall introduced a summer internship at Johnson & Johnson for an Epidemiology UX/Web Design Intern. This internship provides a unique opportunity to merge design principles with epidemiological research, contributing to the advancement of real-world evidence applications. In this role, you will have the opportunity to blend your passion for user experience (UX) and web design with the field of epidemiology, contributing to impactful projects that enhance our ability to derive insights from health data. More information and an application link are available here.
Slides
Video Presentation
 The March 26 community call featured a session on recent publications from the OHDSI community. OHDSI collaborators have published more than 600 studies related to the OMOP CDM and/or OHDSI tools or methods, and we welcomed five lead authors to present their recent publications.
The March 26 community call featured a session on recent publications from the OHDSI community. OHDSI collaborators have published more than 600 studies related to the OMOP CDM and/or OHDSI tools or methods, and we welcomed five lead authors to present their recent publications.
1) Tathagata Bhattacharjee • University of London
INSPIRE datahub: a pan-African integrated suite of services for harmonising longitudinal population health data using OHDSI tools • Frontiers in Digital Health
2) Sulev Reisberg • University of Tartu
Transforming Estonian health data to the Observational Medical Outcomes Partnership (OMOP) Common Data Model: lessons learned • JAMIA Open
3) Fan Bu • University of Michigan
Bayesian safety surveillance with adaptive bias correction • Statistics in Medicine
4) Jen Wooyeon Park • Johns Hopkins University
Development of Medical Imaging Data Standardization for Imaging-Based Observational Research: OMOP Common Data Model Extension • Journal of Imaging Informatics in Medicine
5) Christian Reich • Odysseus
OHDSI Standardized Vocabularies—a large-scale centralized reference ontology for international data harmonization • JAMIA
Community Updates
• Congratulations to the team of Markus Haug, Marek Oja, Maarja Pajusalu, Kerli Mooses, Sulev Reisberg, Jaak Vilo, Antonio Fernández Giménez, Thomas Falconer, Ana Danilović, Filip Maljkovic, Dalia Dawoud, and Raivo Kolde on the publication of Markov modeling for cost-effectiveness using federated health data network in JAMIA.
• Congratulations to the team of Philippe Mortier, Franco Amigo, Madhav Bhargav, Susana Conde, Montse Ferrer, Oskar Flygare, Busenur Kizilaslan, Laura Latorre Moreno, Angela Leis, Miguel Angel Mayer, Víctor Pérez-Sola, Ana Portillo-Van Diest, Juan Manuel Ramírez-Anguita, Ferran Sanz, Gemma Vilagut, Jordi Alonso, Lars Mehlum, Ella Arensman, Johan Bjureberg, Manuel Pastor and Ping Qin on the publication of Developing a clinical decision support system software prototype that assists in the management of patients with self-harm in the emergency department: protocol of the PERMANENS project in BMC Psychiatry.
• The third annual OHDSI DevCon will be held virtually on Friday, April 26, from 9 am-3 pm ET. Join leaders from our Open-Source Community for a day to both welcome and inform both new and veteran developers within the OHDSI Community. More details on the agenda will be posted when available.
• The 2024 OHDSI Global Symposium will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, NJ. The tentative symposium format will feature tutorials on Oct. 22, plenaries and the collaborator showcase on Oct. 23, and workgroup activities on Oct. 24. Registration has not opened yet.
• Registration is now OPEN for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
• Atif Adam announced an opportunity for collaboration around a new network study focusing on Acute ST-Elevation Myocardial Infarction (STEMI). The study intends to deepen the understanding of STEMI patients’ characteristics and identify incidence rates across multiple real-world data datasets. More details and information on how to get involved are available within the forum post.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Synthesizing Evidence for Rare Events: a Novel Zero-Inflated Bivariate Model to Integrate Studies with Double-Zero Outcomes (Lu Li)
Tuesday — Active Safety Surveillance Using Real-world Evidence (ASSURE): An application of the Strategus package (Kevin Haynes)
Wednesday — Patient’s outcomes after endoscopic retrograde cholangiopancreatography (ERCP) using reprocessed duodenoscope accessories: a descriptive study using real-world data (Jessica Mayumi Maruyama)
Thursday — Does COVID-19 Increase Racial/Ethnic Differences in Prevalence of Post-acute Sequelae of SARS-CoV-2 infection (PASC) in Children and Adolescents? an EHR-Based Cohort from the RECOVER Program (Dazheng Zhang)
Friday — Eye Care and Vision Research Workgroup: First Year Update (Michelle Hribar)
Job Openings
• Alex Asiimwe shared an opening for a Director, RWE – Data Science at Gilead. As a RWE Data Scientist (OMOP/OHDSI), you will play a crucial role in designing, implementing, and maintaining healthcare data solutions using the OHDSI framework. This position offers an exciting opportunity to contribute to advancements in health informatics and research.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
• Nathan Hall introduced a summer internship at Johnson & Johnson for an Epidemiology UX/Web Design Intern. This internship provides a unique opportunity to merge design principles with epidemiological research, contributing to the advancement of real-world evidence applications. In this role, you will have the opportunity to blend your passion for user experience (UX) and web design with the field of epidemiology, contributing to impactful projects that enhance our ability to derive insights from health data. More information and an application link are available here.
Slides
Bhattacharjee | Reisberg | Bu | Park | Reich | Community Updates
Videos
INSPIRE datahub: a pan-African integrated suite of services for harmonising longitudinal population health data using OHDSI tools (Bhattacharjee)
Transforming Estonian health data to the Observational Medical Outcomes Partnership (OMOP) Common Data Model: lessons learned (Reisberg)
Bayesian safety surveillance with adaptive bias correction (Bu)
Development of Medical Imaging Data Standardization for Imaging-Based Observational Research: OMOP Common Data Model Extension (Park)
OHDSI Standardized Vocabularies—a large-scale centralized reference ontology for international data harmonization (Reich)
The March 12 community call featured a session on March Madness and April Olympians. March Madness: Less than two weeks after the latest release of OHDSI Standardized Vocabularies, this session found some of the most interesting and unique concepts for a head-to-head showdown, March Madness style. Exploding head syndrome. Dragon’s Blood Extract. Collision of spacecraft with other spacecraft. And God Only Knows what else. April Olympians: Clair Blacketer provided a brief introduction to April Olympians, a month-long community activity that will focus on CDM and Themis conventions. More information and a registration link are below.
Community Updates
• As mentioned above, Clair Blacketer and Melanie Philofsky will lead a month-long community activity in April that will focus on CDM and Themis conventions. There will be five goals of this event:
– Identify all currently ratified CDM and THEMIS conventions for every CDM table and field
– Write clear documentation for each THEMIS convention
– Establish a repository for THEMIS conventions
– Update the CDM documentation to link to relevant THEMIS repository entries
– Create CDM documentation related to expansion module efforts around the community
If you would like to participate in this event, please fill out this form.
• The 2024 OHDSI Global Symposium will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, NJ. The tentative symposium format will feature tutorials on Oct. 22, plenaries and the collaborator showcase on Oct. 23, and workgroup activities on Oct. 24. Registration has not opened yet.
• Registration is now OPEN for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
• Evanette Burrows shared an update about a new release to the ETL-Synthea package v2.0.0 that went live on Feb. 26. The package has been expanded to support current versions of Synthea (v3.1 and v3.2) and has a handful of other improvements and contributions from the community. Full release details are available here.
• Atif Adam announced an opportunity for collaboration around a new network study focusing on Acute ST-Elevation Myocardial Infarction (STEMI). The study intends to deepen the understanding of STEMI patients’ characteristics and identify incidence rates across multiple real-world data datasets. More details and information on how to get involved are available within the forum post.
• James Weaver, an Associate Director of Observational Health Data Analytics at Janssen Research and Development, will speak during a panel session on Current Approaches for Distributed Analysis on Thursday, March 14 (1 pm ET) during a Health Data Research Network Canada event. This will be a virtual conversation; more information and a registration link are available here.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Bladder cancer – a quality benchmark utilizing FHIR and OMOP (Andries Clinckaert)
Tuesday — Using MONAI Pre-Trained Models for Colorectal Tissue Type Phenotyping: A Feasibility Study to Integrate Deep Learning Model Results using the Medical Extension OMOP CDM (Shijia Zhang)
Wednesday — A tool for empirically identifying and reviewing candidate comparators for Pharmacoepidemiological studies (Justin Bohn)
Thursday — The necessity of validity diagnostics when drawing causal inferences from observational data (James Weaver)
Friday — Identification of HIV positive individuals across multiple datasets (Craig Mayer)
Job Openings
• Alex Asiimwe shared an opening for a Director, RWE – Data Science at Gilead. As a RWE Data Scientist (OMOP/OHDSI), you will play a crucial role in designing, implementing, and maintaining healthcare data solutions using the OHDSI framework. This position offers an exciting opportunity to contribute to advancements in health informatics and research.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
• Nathan Hall introduced a summer internship at Johnson & Johnson for an Epidemiology UX/Web Design Intern. This internship provides a unique opportunity to merge design principles with epidemiological research, contributing to the advancement of real-world evidence applications. In this role, you will have the opportunity to blend your passion for user experience (UX) and web design with the field of epidemiology, contributing to impactful projects that enhance our ability to derive insights from health data. More information and an application link are available here.
Slides
April Olympians | Community Updates
Videos
March Madness
April Olympians
 The March 5 community call focused on the latest vocabulary release, which was shared 29Feb2024, as well as a brief wrap-up discussion around Phenotype Phebruary 2024. The main session will be driven by leaders from our vocabulary team:
The March 5 community call focused on the latest vocabulary release, which was shared 29Feb2024, as well as a brief wrap-up discussion around Phenotype Phebruary 2024. The main session will be driven by leaders from our vocabulary team:
- Alexander Davydov, Director, Lead of Medical Ontologies • Odysseus Data Services
- Oleg Zhuk, Vocabulary Technical Lead • Odysseus Data Services
- Anna Ostropolets, Associate Director, Observational Health Data Analytics • Janssen Research and Development
Community Updates
• Congratulations to the team of Aniek F Markus, Peter R Rijnbeek, Jan A Kors, Edward Burn, Talita Duarte-Salles, Markus Haug, Chungsoo Kim, Raivo Kolde, Youngsoo Lee, Hae-Sim Park, Rae Woong Park, Daniel Prieto-Alhambra, Carlen Reyes, Jerry A Krishnan, Guy G Brusselle, and Katia MC Verhamme on the publication of Real-world treatment trajectories of adults with newly diagnosed asthma or COPD in BMJ Open Respiratory Research.
• Congratulations to the team of Behzad Naderalvojoud, Catherine Curtin, Chen Yanover, Tal El-Hay, Byungjin Choi, Rae Woong Park, Javier Gracia Tabuenca, Mary Pat Reeve, Thomas Falconer, Keith Humphreys, Steven M Asch, and Tina Hernandez-Boussard on the publication of Towards global model generalizability: independent cross-site feature evaluation for patient-level risk prediction models using the OHDSI network in JAMIA.
• Congratulations to the team of Star Liu, Asieh Golozar, Nathan Buesgens, Jody-Ann McLeggon, Adam Black, and Paul Nagy on the publication of A framework for understanding an open scientific community using automated harvesting of public artifacts in JAMIA Open.
• Congratulations to the team of Yi Chai, Kenneth K. C. Man, Hao Luo, Carmen Olga Torre, Yun Kwok Wing, Joseph F. Hayes, David P. J. Osborn, Wing Chung Chang, Xiaoyu Lin, Can Yin, Esther W. Chan, Ivan C. H. Lam, Stephen Fortin, David M. Kern, Dong Yun Lee, Rae Woong Park, Jae-Won Jang, Jing Li, Sarah Seager, Wallis C. Y. Lau, and Ian C. K. Wong on the publication of Incidence of mental health diagnoses during the COVID-19 pandemic: a multinational network study in Epidemiology and Psychiatric Sciences.
• Congratulations to the team of Quentin Marcou, Laure Berti-Equille, and Noël Novelli on the publication of Creating a computer assisted ICD coding system: Performance metric choice and use of the ICD hierarchy in the Journal of Biomedical Informatics.
• The 2024 OHDSI Global Symposium will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, NJ. The tentative symposium format will feature tutorials on Oct. 22, plenaries and the collaborator showcase on Oct. 23, and workgroup activities on Oct. 24. Registration has not opened yet.
• Registration is now OPEN for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
• Ross Williams is a scientific researcher working in the group of Dr. Peter Rijnbeek at Erasmus MC, where he is part of the Health Data Science group. His main focus is creating tools and analysis methods to develop personalised medical risk prediction. His specific areas of interest are on the external validation of prediction models, net benefit assessment and techniques for temporal health data analysis. He co-leads both the Patient Level Prediction workgroup and the Early-Stage Researcher workgroup. Ross discusses his career journey, how observational data impacts prediction models, the opportunities for junior researchers in OHDSI, and plenty more in the latest edition of the Collaborator Spotlight.
• The latest edition of the OHDSI newsletter is now available. It includes links to all workgroup OKR presentations from last month, as well as updates from Phenotype Phebruary. It also includes the monthly video podcast, 11 February publications, a new Collaborator Spotlight, and more. If you don’t receive the monthly newsletter in your inbox, please subscribe here.
• James Weaver, an Associate Director of Observational Health Data Analytics at Janssen Research and Development, will speak during a panel session on Current Approaches for Distributed Analysis on Thursday, March 14 (1 pm ET) during a Health Data Research Network Canada event. This will be a virtual conversation; more information and a registration link are available here.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Implementing a common data model in ophthalmology: Comparison of general eye examination mapping to standard OMOP concepts across two major EHR systems (Justin Quon)
Tuesday — Measuring Study Potential Through the Use of Data Diagnostics (Clair Blacketer)
Wednesday — Utilizing Graph Embeddings for Multiple Sclerosis Disease Modifying Therapy Adverse Events (Jason Patterson)
Thursday — Comorbidity Co-occurrence in Women with Endometriosis: A Retrospective Matched Cohort Study (Tamar Zelovich)
Friday — Ulysses: Introducing a workflow R package for assisting in the development of OHDSI studies (Martin Lavallee)
Job Openings
• Priya Desai shared an opening for a Biomedical Informatics Data Scientist at Stanford University who will partner with researchers and clinicians to enable effective and efficient use of data and resources available via Stanford’s research clinical data repository (STARR) including the Electronic Health Records in the OMOP Common Data Model, radiology and cardiology imaging data and associated metadata, and new data types as they get integrated along with their databases and respective cohort query tools and interfaces e.g., OHDSI ATLAS.
• Alex Asiimwe shared three openings at Gilead that could be of interest to OHDSI collaborators. There are openings for a Director, RWE – Data Science, a Director, Data Acquisition, and a Senior Director, Head of Data Office. You can learn more about each position and apply using the links above.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
• Nathan Hall introduced a summer internship at Johnson & Johnson for an Epidemiology UX/Web Design Intern. This internship provides a unique opportunity to merge design principles with epidemiological research, contributing to the advancement of real-world evidence applications. In this role, you will have the opportunity to blend your passion for user experience (UX) and web design with the field of epidemiology, contributing to impactful projects that enhance our ability to derive insights from health data. More information and an application link are available here.
• There is an opening for an Epidemiology Graduate Intern at Johnson & Johnson. Among the responsibilities for this remote position are assisting in managing epidemiologic studies and literature reviews to characterize incidence, prevalence, risk factors, and associated comorbidities and treatment patterns for lung, head, and neck cancers across interventional oncology, contributing to the development of protocols for analyzing real-world data cohorts, such as SEER Medicare, and conducting quantitative analyses using both public and private data sources. More information and an application link are available here.
Slides
Vocabulary Release | Community Updates/Phenotype Phebruary
Videos
Vocabulary Release (Alexander Davydov, Oleg Zhuk, Anna Ostropolets)
Phenotype Phebruary (Azza Shoaibi)
Each community call during “Phenotype Phebruary” features a set of Workgroup 2024 Objectives & Key Result (OKR) announcements, as well an update from that week’s Phenotype Phebruary activities and findings. Workgroups that presented during this call were FHIR + OMOP, Health Equity, the Africa Chapter, Electronic Animal Health Records, CDM Vocabulary, Phenotype Development & Evaluation, Dentistry, Medical Imaging, Medical Devices and GIS – Geographic Information System. The Phenotype Phebruary update, led by Evan Minty and Eva-Maria Didden, reflected on findings from the first three weeks, as well as early research and critical questions around the Week 4 phenotype, pulmonary arterial hypertension.
Community Updates
• Congratulations to the team of Christine Mary Hallinan, Roger Ward, Graeme K Hart, Clair Sullivan, Nicole Pratt, Ashley Ng, Daniel Capurro, Anton Van Der Vegt, Siaw-Teng Liaw, Oliver Daly, Blanca Gallego Luxan, David Bunker and Douglas Boyle on the publication of Seamless EMR data access: Integrated governance, digital health and the OMOP-CDM in BMJ Health & Care Informatics.
• The 2024 OHDSI Global Symposium will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, NJ. The tentative symposium format will feature tutorials and workshops on Oct. 22, the main conference on Oct. 23, and workgroup activities on Oct. 24. Registration has not opened yet.
• Registration is now OPEN for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
• Henrik John recently announced a new network study that he is leading with Chungsoo Kim, Jenna Reps and Egill Fridgeirsson on “Deep Learning Comparison.” The aim is to assess the value of deep learning methods over conventional methods for the development of clinical prediction models. The specific diseases under consideration are dementia in individuals over 55, lung cancer in those over 45, and bipolar disorder in patients misdiagnosed with major depressive disorder. If you would like to join this effort, please read this forum post for more information and reach out to the study leads by March 1.
• James Weaver, an Associate Director of Observational Health Data Analytics at Janssen Research and Development, will speak during a panel session on Current Approaches for Distributed Analysis on Thursday, March 14 (1 pm ET) during a Health Data Research Network Canada event. This will be a virtual conversation; more information and a registration link are available here.
• Under the leadership of Azza Shoaibi, Anna Ostropolets, Gowtham Rao and James Weaver, Phenotype Phebruary 2024 focuses on assessing consistency in phenotype definition components, phenotype representation structure, and phenotype validation methods. The month-long activity empowers OHDSI collaborators to engage with each other while advancing the science of phenotyping and gaining education and training around phenotype development and evaluation. You can check out the event homepage here.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Brazilian administrative data for real-world research: a deterministic linkage procedure and OMOP CDM harmonization (Jessica Mayumi Maruyama)
Tuesday — Estimating model performance on external data sources from their summary statistics: a real-world benchmark (Tal El-Hay)
Wednesday — Harmonization of OMOP vaccine-related vocabularies through the Vaccine Ontology (Yuanyi Pan)
Thursday — OHDSI on Databricks: A Complete Guide to Implementing OHDSI on Databricks (John Gresh)
Friday — Antihypertensive medication use in pregnancy: A pilot OHDSI network analysis in electronic health record data (Stephanie Leonard)
Job Openings
• Alex Asiimwe shared three openings at Gilead that could be of interest to OHDSI collaborators. There are openings for a Director, RWE – Data Science, a Director, Data Acquisition, and a Senior Director, Head of Data Office. You can learn more about each position and apply using the links above.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
• Nathan Hall introduced a summer internship at Johnson & Johnson for an Epidemiology UX/Web Design Intern. This internship provides a unique opportunity to merge design principles with epidemiological research, contributing to the advancement of real-world evidence applications. In this role, you will have the opportunity to blend your passion for user experience (UX) and web design with the field of epidemiology, contributing to impactful projects that enhance our ability to derive insights from health data. More information and an application link are available here.
• There is an opening for an Epidemiology Graduate Intern at Johnson & Johnson. Among the responsibilities for this remote position are assisting in managing epidemiologic studies and literature reviews to characterize incidence, prevalence, risk factors, and associated comorbidities and treatment patterns for lung, head, and neck cancers across interventional oncology, contributing to the development of protocols for analyzing real-world data cohorts, such as SEER Medicare, and conducting quantitative analyses using both public and private data sources. More information and an application link are available here.
• The University of North Carolina Chapel Hill CTSA is hiring a Research Informatics Specialist. This is a remote position (US only, with mostly East Coast hours). This person will join a skilled and highly collaborative team of data analysts, software developers, and data scientists within our CTSA. The core purpose of this position is to support the All of Us Center for Linkage and Acquisition of Data project. We are looking for folks with SQL and health care and/or claims data experience, especially OMOP.
• There is an opening for a Data Steward position at the EBMT. Among the responsibilities is the design, implementation and testing of new data collection processes including data collection forms (DCFs) development, as well as the mapping of new items from DCFs to the OMOP CDM.
Slides
Phenotype Phebruary | Workgroup OKRs + Community Updates
Videos
Workgroup Updates (FHIR + OMOP, Health Equity, the Africa Chapter, Electronic Animal Health Records, CDM Vocabulary, Phenotype Development & Evaluation, Dentistry, Medical Imaging, Medical Devices and GIS – Geographic Information System)
Phenotype Phebruary Update #4
Each community call during “Phenotype Phebruary” features a set of Workgroup 2024 Objectives & Key Result (OKR) announcements, as well an update from that week’s Phenotype Phebruary activities and findings. Workgroups that presented during this call were Themis, Healthcare Systems, Generative AI and Foundational Models, Oncology, Vaccine Vocabulary, Patient-Level Prediction (PLP), ATLAS, Open-Source Community, Psychiatry, and Natural Language Processing (NLP). The Phenotype Phebruary update, led by Anna Ostropolets and Jamie Weaver, reflected on findings from the first two weeks, as well as early research and critical questions around the Week 3 phenotype, major depressive disorder.
Community Updates
• Congratulations to the team of Martin Boeker, Daniela Zöller, Romina Blasini, Philipp Macho, Sven Helfer, Max Behrens, Hans-Ulrich Prokosch and Christian Gulden on the publication of Effectiveness of IT-supported patient recruitment: study protocol for an interrupted time series study at ten German university hospitals in Trials.
• Congratulations to the team of Moshe Zisser and Dvir Aran on the publication of Transformer-based time-to-event prediction for chronic kidney disease deterioration in JAMIA.
• The 2024 OHDSI Global Symposium will be held Oct. 22-24 at the Hyatt Regency Hotel in New Brunswick, NJ. The tentative symposium format will feature tutorials and workshops on Oct. 22, the main conference on Oct. 23, and workgroup activities on Oct. 24. Registration has not opened yet.
• Registration is now OPEN for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
• Henrik John recently announced a new network study that he is leading with Chungsoo Kim, Jenna Reps and Egill Fridgeirsson on “Deep Learning Comparison.” The aim is to assess the value of deep learning methods over conventional methods for the development of clinical prediction models. The specific diseases under consideration are dementia in individuals over 55, lung cancer in those over 45, and bipolar disorder in patients misdiagnosed with major depressive disorder. If you would like to join this effort, please read this forum post for more information and reach out to the study leads by March 1.
• James Weaver, an Associate Director of Observational Health Data Analytics at Janssen Research and Development, will speak during a panel session on Current Approaches for Distributed Analysis on Thursday, March 14 (1 pm ET) during a Health Data Research Network Canada event. This will be a virtual conversation; more information and a registration link are available here.
• Under the leadership of Azza Shoaibi, Anna Ostropolets, Gowtham Rao and James Weaver, Phenotype Phebruary 2024 focuses on assessing consistency in phenotype definition components, phenotype representation structure, and phenotype validation methods. The month-long activity empowers OHDSI collaborators to engage with each other while advancing the science of phenotyping and gaining education and training around phenotype development and evaluation. You can check out the event homepage here.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Challenges and opportunities in adopting OMOP-CDM in Brazilian healthcare: a report from Hospital Israelita Albert Einstein (Maria Abrahao)
Tuesday — Developing a pregnancy algorithm in ATLAS: Applying start date offset (Rupa Makadia)
Wednesday — Creating parsimonious patient-level prediction models using feature selection (Aniek Markus)
Thursday — From Complexity to Clarity: Reproducible and Scalable Phenotype Development and application of LLM in a support role (Asieh Golozar)
Friday — Establishing and Operating the OHDSI Dentistry Workgroup: A Model for Other Disciplines (Danielle Boyce)
Job Openings
• Alex Asiimwe shared three openings at Gilead that could be of interest to OHDSI collaborators. There are openings for a Director, RWE – Data Science, a Director, Data Acquisition, and a Senior Director, Head of Data Office. You can learn more about each position and apply using the links above.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
• Nathan Hall introduced a summer internship at Johnson & Johnson for an Epidemiology UX/Web Design Intern. This internship provides a unique opportunity to merge design principles with epidemiological research, contributing to the advancement of real-world evidence applications. In this role, you will have the opportunity to blend your passion for user experience (UX) and web design with the field of epidemiology, contributing to impactful projects that enhance our ability to derive insights from health data. More information and an application link are available here.
• There is an opening for an Epidemiology Graduate Intern at Johnson & Johnson. Among the responsibilities for this remote position are assisting in managing epidemiologic studies and literature reviews to characterize incidence, prevalence, risk factors, and associated comorbidities and treatment patterns for lung, head, and neck cancers across interventional oncology, contributing to the development of protocols for analyzing real-world data cohorts, such as SEER Medicare, and conducting quantitative analyses using both public and private data sources. More information and an application link are available here.
• The University of North Carolina Chapel Hill CTSA is hiring a Research Informatics Specialist. This is a remote position (US only, with mostly East Coast hours). This person will join a skilled and highly collaborative team of data analysts, software developers, and data scientists within our CTSA. The core purpose of this position is to support the All of Us Center for Linkage and Acquisition of Data project. We are looking for folks with SQL and health care and/or claims data experience, especially OMOP.
• There is an opening for a Data Steward position at the EBMT. Among the responsibilities is the design, implementation and testing of new data collection processes including data collection forms (DCFs) development, as well as the mapping of new items from DCFs to the OMOP CDM.
Slides
Phenotype Phebruary | Workgroup OKRs + Community Updates
Videos
Workgroup Updates (Themis, Healthcare Systems, Generative AI and Analytics in Healthcare (GAIA), Oncology, Vaccine Vocabulary, Patient-Level Prediction, ATLAS, Open Source Community, Psychiatry, and NLP)
Phenotype Phebruary Update #3
Each community call during “Phenotype Phebruary” features a set of Workgroup 2024 Objectives & Key Result (OKR) announcements, as well an update from that week’s Phenotype Phebruary activities and findings. Workgroups that presented during this call were Common Data Model, Network Data Quality, Asia-Pacific (APAC), Industry, Eye Care & Vision Research, and Surgery & Perioperative Medicine. The Phenotype Phebruary update focused on the findings from the Week 1 phenotype, Alzheimer’s Disease, as well as early research and critical questions around the Week 2 phenotype, non-small cell and small cell lung cancer.
Community Updates
• Congratulations to the team of Xinyuan Zhang, Yixue Feng, Fang Li, Jin Ding, Danyal Tahseen, Ezekiel Hinojosa, Yong Chen, and Cui Tao on the publication of Evaluating MedDRA-to-ICD terminology mappings in BMC Medical Informatics and Decision Making.
• Congratulations to the team of Tathagata Bhattacharjee, Sylvia Kiwuwa-Muyingo, Chifundo Kanjala, Molulaqhooa L Maoyi, David Amadi, Michael Ochola, Damazo Kadengye, Arofan Gregory, Agnes Kiragga, Amelia Taylor, Jay Greenfield, Emma Slaymaker, Jim Todd, and the INSPIRE Network on the publication of INSPIRE datahub: a pan-African integrated suite of services for harmonising longitudinal population health data using OHDSI tools in Frontiers in Digital Health.
• Registration is now OPEN for the 2024 OHDSI Europe Symposium, which will be held June 1-3 in Rotterdam, Netherlands. There will be tutorials and workshops June 1-2 at the Erasmus University Medical Center, and the main conference will be held Monday, June 3, on the Steam Ship Rotterdam. Please visit the event homepage for more information and registration details.
• If you are interested in joining the Scientific Review Committee for the 2024 Global Symposium, you can sign up now. The first meeting for the Scientific Review Committee will be held March 7.
• Henrik John recently announced a new network study that he is leading with Chungsoo Kim, Jenna Reps and Egill Fridgeirsson on “Deep Learning Comparison.” The aim is to assess the value of deep learning methods over conventional methods for the development of clinical prediction models. The specific diseases under consideration are dementia in individuals over 55, lung cancer in those over 45, and bipolar disorder in patients misdiagnosed with major depressive disorder. If you would like to join this effort, please read this forum post for more information and reach out to the study leads by March 1.
• James Weaver, an Associate Director of Observational Health Data Analytics at Janssen Research and Development, will speak during a panel session on Current Approaches for Distributed Analysis on Thursday, March 14 (1 pm ET) during a Health Data Research Network Canada event. This will be a virtual conversation; more information and a registration link are available here.
• Under the leadership of Azza Shoaibi, Anna Ostropolets, Gowtham Rao and James Weaver, Phenotype Phebruary 2024 focuses on assessing consistency in phenotype definition components, phenotype representation structure, and phenotype validation methods. The month-long activity empowers OHDSI collaborators to engage with each other while advancing the science of phenotyping and gaining education and training around phenotype development and evaluation. You can check out the event homepage here.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Toward a General-Purpose Geography-Focused OHDSI Infrastructure (Kyle Zollo-Venecek)
Tuesday — The Development and Validation of an Individual-Level Socioeconomic Deprivation Index (ISDI) with OMOP in the NIH’s All of Us Data Network (Nripendra Acharya)
Wednesday — Incorporating measurement values into patient-level prediction with missing entries: a feasibility study (Xiaoyu Wang)
Thursday — The Use of the Julia Programming Language for Global Health Informatics and Observational Health Research (Jacob Zelko)
Friday — Analyzing a Tabloid Headline with Real-World Data: A Summer Intern’s Investigation (Delia Harms)
Job Openings
• Alex Asiimwe shared three openings at Gilead that could be of interest to OHDSI collaborators. There are openings for a Director, RWE – Data Science, a Director, Data Acquisition, and a Senior Director, Head of Data Office. You can learn more about each position and apply using the links above.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
• Nathan Hall introduced a summer internship at Johnson & Johnson for an Epidemiology UX/Web Design Intern. This internship provides a unique opportunity to merge design principles with epidemiological research, contributing to the advancement of real-world evidence applications. In this role, you will have the opportunity to blend your passion for user experience (UX) and web design with the field of epidemiology, contributing to impactful projects that enhance our ability to derive insights from health data. More information and an application link are available here.
• The University of North Carolina Chapel Hill CTSA is hiring a Research Informatics Specialist. This is a remote position (US only, with mostly East Coast hours). This person will join a skilled and highly collaborative team of data analysts, software developers, and data scientists within our CTSA. The core purpose of this position is to support the All of Us Center for Linkage and Acquisition of Data project. We are looking for folks with SQL and health care and/or claims data experience, especially OMOP.
• There is an opening for a Data Steward position at the EBMT. Among the responsibilities is the design, implementation and testing of new data collection processes including data collection forms (DCFs) development, as well as the mapping of new items from DCFs to the OMOP CDM.
Slides
Phenotype Phebruary Update | Workgroup OKRs + Community Updates
Videos
Workgroup Updates (Common Data Model, Network Data Quality, Asia-Pacific (APAC), Industry, Eye Care & Vision Research, and Surgery & Perioperative Medicine)
Phenotype Phebruary Update #2
Each community call during “Phenotype Phebruary” features a set of Workgroup 2024 Objectives & Key Result (OKR) announcements, as well an update from that week’s Phenotype Phebruary activities and findings; the Week 1 phenotype focus was Alzheimer’s Disease. Workgroups that presented during this call were Methods Research, HADES, Perinatal and Reproductive Health, Registry, and the Steering Group.
Community Updates
• If you are interested in joining the Scientific Review Committee for the 2024 Global Symposium, you can sign up now. The first meeting for the Scientific Review Committee will be held March 7.
• Collaborators from both the Columbia University Department of Biomedical Informatics and the Johnson & Johnson Observational Health Data Analytics held a three-day studyathon this past weekend with a focus on women’s health initiatives, specifically endometriosis and polycystic ovary syndrome.
• Kerry Goetz is the Associate Director for the National Eye Institute’s Office of Data Science and Health Informatics at the US National Institutes of Health. In this capacity she is responsible for advancing data management and sharing strategies to make NEI data FAIR (Fully AI-Ready & Findable, Accessible, Interoperable, and Reusable). Kerry co-leads the Eye Care and Vision Research Observational Health Data Sciences and Informatics Working Group. She discusses her career journey, evidence gaps around vision research, how OHDSI impacts her PhD journey, and more in the latest collaborator spotlight.
• The latest edition of The Journey newsletter is now available. It includes details on Phenotype Phebruary, reflections on where OHDSI can go together in 2024, the latest OHDSI videocast, and more community updates. It also includes links to 17 published studies that came out of the OHDSI community in January. If you don’t receive the monthly newsletter in your email, you can subscribe here.
• February community calls will focus on both Phenotype Phebruary updates and 2024 OKR announcements by our various workgroups. All workgroup leads/representatives should sign up for one of the upcoming community calls using this link.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — FinOMOP – a population-based data network (Javier Gracia-Tabuenca)
Tuesday — Operational Definition of Adrenal diseases: Enhancing Precision and Reproducibility in Observational Data (Suhyun Kim)
Wednesday — Impact of concomitant use of proton pump inhibitors and clopidogrel on cardiovascular adverse outcomes – A multicenter study using common data model (Seonji Kim)
Thursday — Leveraging the OMOP Common Data Model to Support Distributed Health Equity Research (Sarah Gasman)
Friday — Validating a clinical informatics consulting service using negative control reference sets (Michael Jackson)
HADES Development Updates
• Martijn Schuemie announced the release of CohortMethod 5.2.1. The most important changes are updating the Capr function calls in the vignettes (the old code was no longer working), and CohortMethod now asks if you want to delete old files when you call runCmAnalyses() using an existing folder but different analysis settings than before.
• Ger Inberg announced the release of FeatureExtraction 3.4.0. It contains mainly bugfixes and furthermore the ‘cohortId’ argument in exported functions has been deprecated, one should use ‘cohortIds’ instead.
Job Openings
• Alex Asiimwe shared three openings at Gilead that could be of interest to OHDSI collaborators. There are openings for a Director, RWE – Data Science, a Director, Data Acquisition, and a Senior Director, Head of Data Office. You can learn more about each position and apply using the links above.
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
• Nathan Hall introduced a summer internship at Johnson & Johnson for an Epidemiology UX/Web Design Intern. This internship provides a unique opportunity to merge design principles with epidemiological research, contributing to the advancement of real-world evidence applications. In this role, you will have the opportunity to blend your passion for user experience (UX) and web design with the field of epidemiology, contributing to impactful projects that enhance our ability to derive insights from health data. More information and an application link are available here.
• The University of North Carolina Chapel Hill CTSA is hiring a Research Informatics Specialist. This is a remote position (US only, with mostly East Coast hours). This person will join a skilled and highly collaborative team of data analysts, software developers, and data scientists within our CTSA. The core purpose of this position is to support the All of Us Center for Linkage and Acquisition of Data project. We are looking for folks with SQL and health care and/or claims data experience, especially OMOP.
• There is an opening for a Data Steward position at the EBMT. Among the responsibilities is the design, implementation and testing of new data collection processes including data collection forms (DCFs) development, as well as the mapping of new items from DCFs to the OMOP CDM.
Slides
Phenotype Phebruary Update | Workgroup OKRs + Community Updates
Videos
Workgroup Updates (Methods Research, HADES, Perinatal and Reproductive Health, Registry, Steering Group)
Phenotype Phebruary Update #1
 The third installment of Phenotype Phebruary is approaching, and the leadership team provided an overview of the initiative, its importance in research, and how this version of Phenotype Phebruary will take place in the OHDSI community. This talk also included a “Phenotype 101” session, as well as a community vote on four phenotypes to be focused on during the month. The selections were Alzheimer’s, pulmonary hypertension, major depression disorder and prostate cancer. This session was led by:
The third installment of Phenotype Phebruary is approaching, and the leadership team provided an overview of the initiative, its importance in research, and how this version of Phenotype Phebruary will take place in the OHDSI community. This talk also included a “Phenotype 101” session, as well as a community vote on four phenotypes to be focused on during the month. The selections were Alzheimer’s, pulmonary hypertension, major depression disorder and prostate cancer. This session was led by:
• Azza Shoaibi – Director, Observational Health Data Analytics, Janssen Research and Development
• Jamie Weaver – Associate Director, Observational Health Data Analytics, Janssen Research and Development
• Anna Ostropolets – Associate Director, Observational Health Data Analytics, Janssen Research and Development
Patrick Ryan and Asieh Golozar also provided comments. You can sign up to take part in Phenotype Phebruary, and you can follow updates via the OHDSI forums.
Community Updates
• Congratulations to the team of Soobeen Seol, Jimyung Park, Chungsoo Kim, Dong Yun Lee, and Rae Woong Park on the publication of RHEA: Real-World Observational Health Data Exploration Application in Volume 310 of Studies in Health Technology and Informatics.
• Congratulations to the team of Sujin Gan, Chungsoo Kim, Dong Yun Lee, and Rae Woong Park on the publication of Prediction Models for Readmission Using Home Healthcare Notes and OMOP-CDM in Volume 310 of Studies in Health Technology and Informatics.
• Congratulations to the team of Scott L. DuVall, Craig G. Parker, Amanda R. Shields, Patrick R. Alba, Julie A. Lynch, Michael E. Matheny, and Aaron W.C. Kamauu on the publication of Toward Real-World Reproducibility: Verifying Value Sets for Clinical Research in Volume 310 of Studies in Health Technology and Informatics.
• Congratulations to the team of Piper Ranallo, Bronwyn Southwell, Christopher Tignanelli, Steven G. Johnson, Richard Krueger, Tess Sevareid-Groth, Adam Carvel, and Genevieve B. Melton on the publication of Promoting Learning Health System Cycles by Optimizing EHR Data Clinical Concept Encoding Processes in Volume 310 of Studies in Health Technology and Informatics.
• Congratulations to the team of ChulHyoung Park, Sang Jun Park, Da Yun Lee, Seng Chan You, Kihwang Lee, and Rae Woong Park on the publication of Multi-Institutional Collaborative Research Using Ophthalmic Medical Image Data Standardized by Radiology Common Data Model (R-CDM) in Volume 310 of Studies in Health Technology and Informatics.
• Congratulations to the team of Seol Whan Oh, Soo Jeong Ko, Yun Seon Im, Surin Jung, Bo Yeon Choi, Jae Yoon Kim, Sunghyeon Park, Wona Choi, and In Young Choi on the publication of Development of Integrated Data Quality Management System for Observational Medical Outcomes Partnership Common Data Model in Volume 310 of Studies in Health Technology and Informatics.
• Congratulations to the team of Martijn Schuemie, Jenna Reps, Adam Black, Frank Defalco, Lee Evans, Egill Fridgeirsson, James P. Gilbert, Chris Knoll, Martin Lavallee, Gowtham A. Rao, Peter Rijnbeek, Katy Sadowski, Anthony Sena, Joel Swerdel, Ross D. Williams, and Marc Suchard on the publication of Health-Analytics Data to Evidence Suite (HADES): Open-Source Software for Observational Research in Volume 310 of Studies in Health Technology and Informatics.
• Congratulations to the team of Jiyun Cha, Eun Kyoung Ahn, Young-Heum Yoon, and Man Young Park on the publication of Feasibility of Applying the OMOP Common Data Model to Traditional Eastern Asian Medicine Dataset in Volume 310 of Studies in Health Technology and Informatics.
• Congratulations to the team of Najia Ahmadi, Quang Vu Nguyen, Martin Sedlmayr, and Markus Wolfien on the publication of A comparative patient-level prediction study in OMOP CDM: applicative potential and insights from synthetic data in Scientific Reports.
• February community calls will focus on both Phenotype Phebruary updates and 2024 OKR announcements by our various workgroups. All workgroup leads/representatives should sign up for one of the upcoming community calls using this link.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Transforming the Optum® Enriched Oncology module to OMOP CDM (Dmitry Dymshyts)
Tuesday — Community Contribution to the OHDSI Vocabularies: moving towards collaborative shared resource (Oleg Zhuk)
Wednesday — Evaluating confounding adjustment when sample size is small (Martijn Schuemie)
Thursday — Using a Continuous Quality Improvement (CQI) Approach for Gap Analysis of OHDSI/ATLAS as An Enterprise Self-Service Analytics Platform by Academic Medical Centers (Selvin Soby)
Friday — Risk of aortic aneurysm or dissection following exposure to fluoroquinolone antibiotics (Jack Jenetzki)
HADES Development Updates
• Martijn Schuemie announced the release of SelfControlledCaseSeries 5.1.1. This contains a fix of a minor bug introduced in 5.1.0.
Job Openings
• Linying Zhang shared openings for both a Postdoc and a Senior Data Analyst at the Washington University School of Medicine in St. Louis. Successful candidates will work on causal machine learning and responsible AI for reliable real-world evidence generation. Interested applicants should send a CV and cover letter to linyingz@wustl.edu.
• The University of North Carolina Chapel Hill CTSA is hiring a Research Informatics Specialist. This is a remote position (US only, with mostly East Coast hours). This person will join a skilled and highly collaborative team of data analysts, software developers, and data scientists within our CTSA. The core purpose of this position is to support the All of Us Center for Linkage and Acquisition of Data project. We are looking for folks with SQL and health care and/or claims data experience, especially OMOP.
• There is an opening for a Data Steward position at the EBMT. Among the responsibilities is the design, implementation and testing of new data collection processes including data collection forms (DCFs) development, as well as the mapping of new items from DCFs to the OMOP CDM.
Slides
Introduction to Phenotype Phebruary | Community Updates
Video Presentation
 The Jan. 23 community call looked at the recent UK Study-A-Thon, held last November at Saint Hilda’s College. The event focused on the use of fluoroquinolones across geographies and over time, as well as on the epidemiology and characterization of rectal prolapse and rectopexy. At the end of an intense week, the team generated three draft manuscripts almost ready for submission, and at least four conference abstracts were in the making.
The Jan. 23 community call looked at the recent UK Study-A-Thon, held last November at Saint Hilda’s College. The event focused on the use of fluoroquinolones across geographies and over time, as well as on the epidemiology and characterization of rectal prolapse and rectopexy. At the end of an intense week, the team generated three draft manuscripts almost ready for submission, and at least four conference abstracts were in the making.
Community Updates
• Congratulations to the team of Xu Zuo, Yujia Zhou, Jon Duke, George Hripcsak, Nigam Shah, Juan Banda, Ruth Reeves, Timothy Miller, Lemuel Waitman, Karthik Natarajan, and Hua Xu on the publication of Standardizing Multi-site Clinical Note Titles to LOINC Document Ontology: A Transformer-based Approach in the AMIA Annual Symposium Proceedings.
• Congratulations to the team of Huzaifa Khan, Abu Saleh Mohammad Mosa, Vyshnavi Paka, Md Kamruz Zaman Rana, Vasanthi Mandhadi, Soliman Islam, Hua Xu, James C McClay, Sraboni Sarker, Praveen Rao, and Lemuel Waitman on the publication of Mapping Clinical Documents to the Logical Observation Identifiers, Names and Codes (LOINC) Document Ontology using Electronic Health Record Systems Structured Metadata in the AMIA Annual Symposium Proceedings.
• Congratulations to the team of Joel Swerdel and Mitchell Conover on the publication of Comparing broad and narrow phenotype algorithms: differences in performance characteristics and immortal time incurred in the Journal of Pharmacy & Pharmaceutical Sciences.
• Registration is open for the 2024 Oxford Summer School: Real world evidence using the OMOP Common Data Model, which will be held June 17-21, 2024 at the University of Oxford. The Real World Evidence Summer School will provide participants with the tools and concepts necessary to plan and execute Real World Evidence studies, with a focus on the use of the OMOP common data model. Learn more about the program here.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Mapping of Critical Care EHR Flowsheet data to the OMOP CDM via SSSOM (Polina Talapova)
Tuesday — Paving the way to estimate daily dose in OMOP CDM for Drug Utilisation Studies in DARWIN EU® (Theresa Burkard)
Wednesday — Generating Synthetic Electronic Health Records in OMOP using GPT (Chao Peng)
Thursday — Comparing concepts extracted from clinical Dutch text to conditions in the structured data (Tom Seinen)
Friday — Finding a constrained number of predictor phenotypes for multiple outcome prediction (Jenna Repa)
Job Openings
• The University of North Carolina Chapel Hill CTSA is hiring a Research Informatics Specialist. This is a remote position (US only, with mostly East Coast hours). This person will join a skilled and highly collaborative team of data analysts, software developers, and data scientists within our CTSA. The core purpose of this position is to support the All of Us Center for Linkage and Acquisition of Data project. We are looking for folks with SQL and health care and/or claims data experience, especially OMOP.
• There is an opening for a Data Steward position at the EBMT. Among the responsibilities is the design, implementation and testing of new data collection processes including data collection forms (DCFs) development, as well as the mapping of new items from DCFs to the OMOP CDM.
Slides
MHRA and the use of RWE | Use of systemic fluoroquinolones in primary care and hospital settings in the UK: a drug utilisation study | Rectopexy & the search for devices | Community Updates
Video Presentation
0:00 – Daniel Prieto-Alhambra – Professor of Pharmaco- and Device Epidemiology, University of Oxford (Introduction)
1:28 – Katherine Donegan – Head of Epidemiology, MHRA (MHRA and the use of RWE)
13:40 – Annika Jodicke – Senior Researcher in Pharmacoepidemiology, University of Oxford (Use of systemic fluoroquinolones in primary care and hospital settings in the UK: a drug utilisation study)
28:52 – Jennifer Lane – NIHR Clinical Lecturer in Trauma and Orthopaedic Surgery, Barts Bone and Joint Health, Queen Mary University of London (Rectopexy & the search for devices)
The Jan. 16 community call featured multiple unrecorded breakout session meant to stimulate future collaboration opportunities throughout the community. The video below highlights the updates shared at the beginning of the call, including a brief presentation on a recent community publication, Real-World Effectiveness of BNT162b2 Against Infection and Severe Diseases in Children and Adolescents.
Community Updates
• Congratulations to the team of Qiong Wu, Jiayi Tong, Bingyu Zhang, Dazheng Zhang, Jiajie Chen, Yuqing Lei, Yiwen Lu, Yudong Wang, Lu Li, Yishan Shen, Jie Xu, L. Charles Bailey, Jiang Bian, Dimitri A. Christakis, Megan L. Fitzgerald, Kathryn Hirabayashi, Ravi Jhaveri, Alka Khaitan, Tianchen Lyu, Suchitra Rao, Hanieh Razzaghi, Hayden T. Schwenk, Fei Wang, Margot I. Gage Witvliet, Eric J. Tchetgen Tchetgen, Jeffrey S. Morris, Christopher B. Forrest, and Yong Chen on the publication of Real-World Effectiveness of BNT162b2 Against Infection and Severe Diseases in Children and Adolescents in Annals of Internal Medicine.
• Congratulations to the team of Martí Català, Núria Mercadé-Besora, Raivo Kolde, Nhung T H Trinh, Elena Roel, Edward Burn, Trishna Rathod-Mistry, Kristin Kostka, Wai Yi Man, Antonella Delmestri, Prof Hedvig M E Nordeng, Anneli Uusküla, Talita Duarte-Salles, Daniel Prieto-Alhambra, and Annika M Jödicke on the publication of The effectiveness of COVID-19 vaccines to prevent long COVID symptoms: staggered cohort study of data from the UK, Spain, and Estonia in The Lancet Respiratory Medicine.
• Anna Ostropolets will lead the next edition of the CBER Best Seminar Seminar series, which will be held Wednesday, Jan. 17, at 11 am ET. Anna will lead a session entitled “KEEPER: Standardized structured data from electronic health records as an alternative to chart review for case adjudication and phenotype evaluation” which will be virtual and available to anybody. More information and the registration link are available here.
• Chungsoo Kim is a PhD candidate in the Department of Biomedical Informatics at Ajou University College of Medicine. Since joining OHDSI in 2019, he has participated in and led several research projects at OHDSI. He currently participates in OHDSI working groups, including PatientLevelPrediction and the APAC group. He also served as a tutorial instructor for the 2019 OHDSI Korea International Symposium. Chungsoo discusses his research focuses, his involvement in the OHDSI community, the growth of OHDSI around the Asia-Pacific region, and plenty more in the latest Collaborator Spotlight.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — Development of psychiatric common data model (P-CDM) leveraging psychiatric scales (Dong Yun Lee)
Tuesday — Integrating large language models and real-world evidence into an automated drug indication taxonomy development workflow (Yilu Fang)
Wednesday — Making OHDSI Tooling accessible to Researchers and Students in a HIPAA Compliant Platform (Hannah Morgan-Cooper)
Thursday — Trend in Prescription Pattern in Heart Failure Medications (Septi Melisa)
Friday — PheMIME: An Interactive Web App and Knowledge Base for Phenome-Wide Multi-Institutional Multimorbidity Analysis (Siwei Zhang)
Job Openings
• There is an opening for a Data Steward position at the EBMT. Among the responsibilities is the design, implementation and testing of new data collection processes including data collection forms (DCFs) development, as well as the mapping of new items from DCFs to the OMOP CDM.
Slides
Video Presentation
 Happy New Year! Patrick Ryan led the first community call of the year for a discussion on what OHDSI can achieve together in 2024. You can watch the recording or check out the slides below.
Happy New Year! Patrick Ryan led the first community call of the year for a discussion on what OHDSI can achieve together in 2024. You can watch the recording or check out the slides below.
Community Updates
• Congratulations to the team of Yoon Jin Choi, Tae Jun Kim, Chang Seok Bang, Yong Kang Lee, Moon Won Lee, Su Youn Nam, Woon Geon Shin, and Seung In Seo on the publication of Changing trends and characteristics of peptic ulcer disease: A multicenter study from 2010 to 2019 in Korea in the World Journal of Gastroenterology.
• Congratulations to the team of Pierre Heudel, Hugo Crochet, Thierry Durand, Philippe Zrounba, and JeanYves Blay on the publication of From data strategy to implementation to advance cancer research and cancer care: A French comprehensive cancer center experience in PLOS Digital Health.
• Congratulations to the team of Cynthia Yang, Egill Fridgeirsson, Jan Kors, Jenna Reps and Peter Rijnbeek on the publication of Impact of random oversampling and random undersampling on the performance of prediction models developed using observational health data in the Journal of Big Data.
• Congratulations to the team of Christian Reich, Anna Ostropolets, Patrick Ryan, Peter Rijnbeek, Martijn Schuemie, Alexander Davydov, Dmitry Dymshyts, and George Hripcsak on the publication of OHDSI Standardized Vocabularies-a large-scale centralized reference ontology for international data harmonization in JAMIA.
• Congratulations to the team of Qiong Wu, Jiayi Tong, Bingyu Zhang, Dazheng Zhang, Jiajie Chen, Yuqing Lei, Yiwen Lu, Yudong Wang, Lu Li, Yishan Shen, Jie Xu, L. Charles Bailey, Jiang Bian, Dimitri A. Christakis, Megan L. Fitzgerald, Kathryn Hirabayashi, Ravi Jhaveri, Alka Khaitan, Tianchen Lyu, Suchitra Rao, Hanieh Razzaghi, Hayden T. Schwenk, Fei Wang, Margot I. Gage Witvliet, Eric J. Tchetgen Tchetgen, Jeffrey S. Morris, Christopher B. Forrest, and Yong Chen on the publication of Real-World Effectiveness of BNT162b2 Against Infection and Severe Diseases in Children and Adolescents in Annals of Internal Medicine.
• Chungsoo Kim is a PhD candidate in the Department of Biomedical Informatics at Ajou University College of Medicine. Since joining OHDSI in 2019, he has participated in and led several research projects at OHDSI. He currently participates in OHDSI working groups, including PatientLevelPrediction and the APAC group. He also served as a tutorial instructor for the 2019 OHDSI Korea International Symposium. Chungsoo discusses his research focuses, his involvement in the OHDSI community, the growth of OHDSI around the Asia-Pacific region, and plenty more in the latest Collaborator Spotlight.
• The latest edition of the OHDSI newsletter is now available, and it includes the December video presentations of the year in review and OHDSI’s first 10 years. It also features community updates, December publications and presentations, and plenty more.
• Research from the 2023 Global Symposium Collaborator Showcase can be viewed on the Global Symposium Showcase page. Research is also being shared daily on OHDSI’s LinkedIn, Twitter/X and Instagram feeds as part of the #OHDSISocialShowcase. Below are posters (with study leads) that are featured this week:
Monday — A Toxin Vocabulary for the OMOP CDM Park (Maksym Trofymenko)
Tuesday — Developing a perinatal expansion table for the OMOP common data model (Alicia Abellan)
Wednesday — External validation using clinical domain knowledge from the SNOMED medical terms hierarchy (Henrik John)
Thursday — Estimating the comparative risk of kidney failure associated with intravitreal anti-vascular endothelial growth factor (anti-VEGF) exposure in patients with blinding diseases (Cindy Cai)
Friday — Characteristics and outcomes of over a million inflammatory bowel disease subjects in seven countries: a multinational cohort study (Chen Yanover)
HADES Development Updates
• Martiijn Schuemie announced the releases of CohortMethod 5.2.0 and EmpiricalCalibration 3.1.2. The CohortMethod release focuses on generalizability, by comparing the population after all adjustments (e.g. PS matching) to the population before all adjustments, and report any characteristics that have changed. The choice of population to consider for generalizability is now driven by your analysis choices. For example, if you use PS matching, (implying ATT), generalizability will be computed for the target (treated) population.
• Egill Fridgeirsson announced the release of DeepPatientLevelPrediction 2.0.3. This bugfix release only includes one change which was necessary because one of my dependencies introduced a breaking change (polars) which broke some functionality in the package.
• Joel Swerdel announced the release of PheValuator 2.2.11. This adds the capability to include multiple visits per subject in the evaluation cohort.
Job Openings
• There is an opening for a Data Steward position at the EBMT. Among the responsibilities is the design, implementation and testing of new data collection processes including data collection forms (DCFs) development, as well as the mapping of new items from DCFs to the OMOP CDM.
Slides
Main Presentation | Community Updates

